OPEN-ACCESS PEER-REVIEWED
REVIEW ARTICLE
Lùcia Carrano1,* and Flavia Marinelli2
1Fondazione Instituto Insubrico Ricerca per la Vita (FIIRV), 21040 Gerenzano (Va) Italy. 2Dipartimento di Biotecnologie e Scienze della Vita, Università degli Studi dell’Insubria, 21100 Varese and “The Protein Factory” Research Center, Politecnico of Milano, ICRM CNR Milano and University of Insubria, Varese, Italy.
Journal of Applied Bioanalysis. Vol.1. No.2. pages 55-67 (2015)
Published 15 April 2015. https://doi.org/10.17145/jab.15.010 | (ISSN 2405-710X)
Correspondence:
*Carrano L . Fondazione Instituto Insubrico Ricerca per la Vita (FIIRV), via Lepetit 34, 21040 Gerenzano (Va), Italy. Phone +39-02964743229; Fax +39-029647432.
Citation:
Carrano L and Marinelli F. The relevance of chemical dereplication in microbial natural product screening. J Appl Bioanal 1(2), 55-67 (2015).
Open-access and Copyright:
©2015 Carrano L and Marinelli F. This article is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Funding/Manuscript writing assistance:
The authors declare that no writing assistance was utilized in the production of this article. Financial and/or material support was received for this research, the financial sources are clearly identified and acknowledged in this article.
Competing interest:
The authors have declared that no competing interest exist.
Article history:
Received: 30 November 2014, Revised 25 January 2015, Accepted 6 March 2015.
Abstract
Microbial products continue to represent one of the most interesting sources for the discovery and development of novel drugs. As a result of the massive screening of microbes since 1950, one of the most important hindrances encountered so far in screening microbial natural products is re-isolation of already discovered bioactive molecules. Thus, identifying known or undesirable compounds from natural product extracts at an early stage, indicated here as chemical dereplication, is a key step in the process, saving resources and speeding up the discovery process of novel drugs. In this mini-review, we highlight the analytical techniques commonly used to evaluate the novelty of microbial metabolites during screening and the advances that have been made in related technologies.
Kewords
dereplication, natural product, microbial extract, LC-MS structure identification
Introduction
Natural products (NPs) continue to play an important role for the discovery of new therapeutic candidates. Over the past 30 years, NPs or their derivatives have accounted for 60% of new anticancer agents and almost 75% of all new antibacterial molecules [1-3]. One hundred NP and NP-derived substances were being evaluated in clinical trials or were being registered at the end of 2013 [4]. NPs have been isolated from many terrestrial and marine organisms, including plants, marine invertebrates, and microorganisms, the latter being the source selected more often for pharmaceutical drug discovery programs. Microorganisms (traditionally actinobacteria and fungi, but more recently cyanobacteria and myxobacteria as well) are one of the most prolific sources among living organisms for the production of bioactive molecules. Exploitation of their specialized (commonly termed secondary) metabolism has guaranteed for decades already the discovery of novel antibiotics and other compounds with unprecedented chemical characteristics and biological properties not existing in screening libraries of synthetic compounds [1,5]. Querying the literature, we previously reported [6,7] that among more than 31600 microbial products discovered from 1900 onwards, ca. 20200 possess some biological activity. Among them 35% were produced from filamentous fungi, 48% from actinomycetes, and 17% from other bacteria. According to Berdy [2] ca. 20000 and 22000 bioactive microbial secondary metabolites had been described in the scientific and patent literature by the end of 2000 and 2002, respectively. About 38% of these molecules are produced by filamentous fungi, whereas the largest group (45%) derives from actinomycetes (7600 metabolites from Streptomyces and 2500 from the so-called rare filamentous actinomycetes). The remaining 17% is produced by other bacteria such as Bacillus, Pseudomonas, myxobacteria, and cyanobacteria. During the past 15 years, we have registered the progressive decline of the pharmaceutical industry’s interest in NP screening, in part because of the emphasis on high-throughput screening (HTS) of synthetic libraries, but also due to a prevailing sentiment nowadays that screening of natural sources is a time-consuming effort with a high risk of re-finding already known compounds. Recent commentaries on the industrial perspectives concerning drug sources have been published by both academic and industrial researchers and can be assessed by interested readers in [1,3,5,8-10]. As a result of the massive screening of soil actinomycetes and fungi since 1950, one of the most important hindrances encountered so far is the re-isolation of already discovered bioactive molecules. According to Baltz [11], if 10000 actinomycetes were screened, 2500 microbial isolates would produce antibiotics and, among those, 2250 would make streptothricin (2×10-1), 125 streptomycin (1×10-2) and 40 tetracycline (4×10-3); the frequency of rediscovering vancomycin would be 1.5×10-5, erythromycin 5×10-6, and daptomycin 1×10-7. This redundancy of known microbial metabolites originating from complex samples is perceived as the root of the technical difficulties that are intrinsic to NP screening. It has been estimated that each gram of soil contains 106-108 bacteria, 104-106 actinomycetes spores, and 102-104 fungal spores [10-11]; the microbial extracts generated from cultivating each single microbial isolate is estimated to contain complex mixture of metabolites, each of them in concentrations ranging from less than 1 mg/l up to 100 mg/l culture [12]. The chemical complexity of these extracts makes the process of purifying bioactive molecules and elucidating the structures extremely slow and demanding. The recent dramatic developments in microbial ecology and genomics, in our understanding of specialized metabolite biosynthesis and regulation, together with the advances in analytical techniques have prompted renewed and increasing interest in NP screening [3,13-14]. The field was reinvigorated after many actinomycete genomes were sequenced since this unveiled that approximately 70% of the products from putative NP-producing gene clusters have not been characterized [3,13]. Current estimates predict that 109 NPs remain to be isolated [13].
Dereplication along the NP screening
For the reasons stated above, the process termed dereplication, i.e. the process of distinguishing those NP extracts that contain known bioactive metabolites from those that contain novel compounds of interest, is a key methodology in NP-based discovery programs nowadays. The term was first introduced in the first CRC Handbook of antibiotic compounds to define the recognition and elimination of already described active molecules [12,15-16]. In this initial definition, the term was mostly related to what we currently call the chemical dereplication process that is preferentially based on mass spectrometry (MS). MS is one of the two predominant experimental analysis techniques for detecting and identifying metabolites and other small molecules, the other being nuclear magnetic resonance (NMR). The most important advantage of MS over NMR is that it is orders of magnitude more sensitive, making it the method of choice for first-pass compound detection and identification in medium- to high-throughput screening applications. Thus, MS coupled with chromatographic separation and combined with UV-visible detection has become the most widely used analytical technique for analyzing complex mixtures such as microbial extracts [8,13,16-20] .
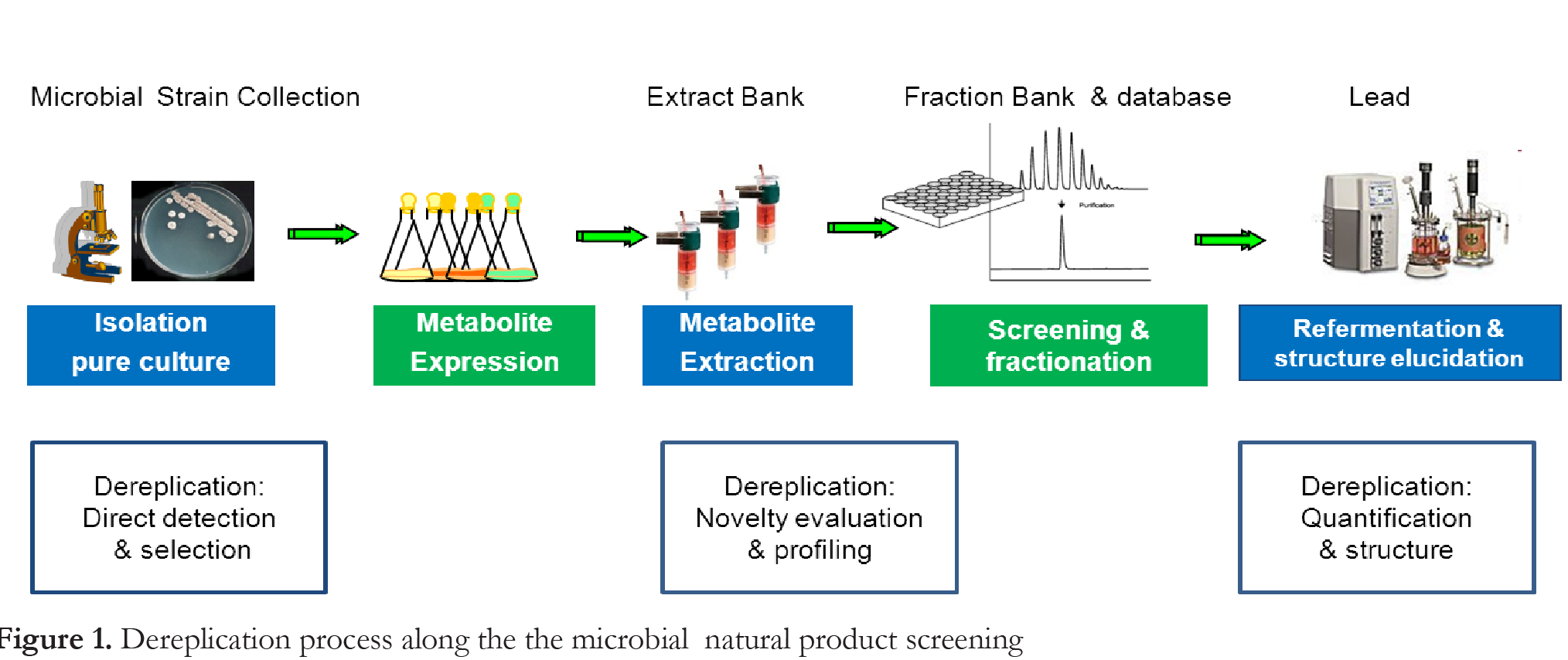
As shown in Figure 1 in simplified form, the term dereplication has been used progressively in NP screening to indicate three different, sequential steps in the discovery process. As the chemical diversity of a microbial extract correlates with the diversity of microbes that it is comprised of, the introduction of novel and unique microbial strains in screening is considered a critical factor for discovering new molecules [6,7]. If large microbial collections are used to generate microbial extracts, redundant bacterial and fungal isolates need to be removed and the number of microbial strains reduced prior to fermentation and subsequent biological and chemical screening [16,19,21]. Then, the term dereplication also started to be used in the sense of eliminating similar microbial strains in the very early stage of NP screening, including microbiological methods such as morphological characterization of colonies grown on several solid media and/or genome analysis by using molecular methods such as partial 16S rDNA sequencing, 16S-ITS restriction fragment length polymorphism (RFLP), and repetitive extragenic palindromic PCR of the BOX DNA element (BOX-PCR) [17,21-26]. This step is indicated as “Direct detection and isolation” in Figure 1.
The critical differentiator of a set of microbial isolates is the presence of a unique NP composition in the extracts they generate through fermentation. A rapid and robust means of identifying the known compounds present in the mixture is crucial to hasten the discovery of novel drugs. Here, chemical dereplication represents the strategy to eliminate known and redundant compounds, indicating which peak in a high-performance liquid chromatographic (HPLC) profile is worthy of being further purified. This is particularly important when biological activity-guided primary screening targets are not selected despite intelligent selection of the microbial sources [16,21,27]. HPLC remains the most versatile technique to efficiently separate specialized metabolites directly from a crude mixture without the need for preparing complex samples. The remarkable improvements in hyphenated analytical methods (i.e. those developed from the coupling of a separation technique and online spectroscopic detection technology) over the last two decades have significantly broadened LC applications in the analysis of NPs [8,13,20]. The most widely used spectroscopic detection methods in this phase (termed “Novelty evaluation and profiling” in Figure 1) are based on MS and and diode array detection (DAD), whose efficacy is supported by increasingly applying bioinformatics methods and developing NP databases (DBs). Using novel MS/MS interface and NMR spectroscopy represent recent improvements [12,18,20,23,28-29]. One of the most critical bottlenecks in NP screening is to structurally elucidate a novel chemical entity that has successfully passed the novelty evaluation step. The identification and quantification of the bioactive compounds in microbial product extracts rely on bioassay-guided fractionation and further purification of the active molecule from new large-scale refermentation of the original microbial producer [20-21,27-29]. During fractionation, NMR and LC-MS are then applied not only to assess the novelty of the compounds, but also to generate the range of spectra necessary to subsequently elucidate the structure of the novel molecules [20-21,29-30]. For this task, the method of choice remains NMR. Consolidated NMR applications for structural elucidation require from ten to hundreds of milligrams of pure compounds (>95% purity), implicating a labor-intensive phase in NP purification and quantification [28-31]. This phase is indicated as “Quantification and Structure” in Figure 1 and needs to be made more efficient in terms of time by making advances in hyphenated and more sensitive NMR methodologies, facilitating the acquisition of MS/MS, and comparing spectra during dereplication. In summary, this paper focuses on the analytical methods developed for the following three steps : (i) direct detection and isolation from microbial colonies, (ii) novelty evaluation and profiling of the active peak in microbial extracts, and (iii) quantification and structure identification of a novel NP (Figure 1).
Direct detection and isolation
Ideally, detecting the specialized metabolites produced by microorganisms directly from the surface of the microbial colonies and on cultivation plates would dramatically speed up the screening process as preparative steps would be eliminated. The emerging new MS technology, combined with sensitive bioassays and an appropriate database of compounds, has prompted several authors to claim that in the postgenomics era, this is possible [12,19,25,30]. In fact, ambient ionization MS methods such as laser ablation electrospray ionization (LAESI, which is a combination of microsampling by a mid-IR laser and efficient ionization of the sampled material by critically charged droplets [32]) or nanospray desorption electrospray ionization (nanoDESI) [33] may be able to directly image the NPs produced by plant leaves, algae, and microorganisms. NanoDESI imaging MS was adapted for in vivo metabolic profiling of living bacterial colonies directly from the Petri dishes and demonstrated the ability to capture a wide variety of molecular classes within a single mass spectrum directly from a live specimen [33-34]. For example, nanoDESI –MS was coupled with alignment of MS data, MS/MS, and molecular networks to dereplicate the antifungal molecules produced by Pseudomonas aeruginosa sp. SH-C52, which protects sugar beet plants from infections by specific soil-borne fungi, identifying thanamycin, a predicted lipopeptide encoded by a non-ribosomal peptide synthetase gene cluster [34]. An increasing number of studies are applying ambient MS technology directly on single colonies in Petri dishes or on paper for analyzing metabolomic pathways, characterizing strains, and identifying novel metabolites [35-37]. Nevertheless, technical problems, such as the capillary clogging in NanoDESI, are currently limiting ambient MS technology from being applied routinely in discovery programs. Different sampling methods were proposed, such as liquid micro-junction surface sampling probe (LMJ-SSP), which is used to perform metabolomic studies on yeast, fungi, and marine bacteria [38]. A screening platform was developed by combining liquid extraction surface analysis (LESA), automated chip-based NanoESI, and high-resolution mass spectrometry (HRMS) or tandem mass spectrometry using an Orbitrap XL [39]: actinobacteria were cultivated on solid agar media; the resulting thiazolyl peptide antibiotics were extracted by organic solvent mixtures from the surface of colonies and analyzed by MS. Future developments in ambient ionization will certainly increase the ability to detect, identify, and dereplicate NPs “in situ” directly at their microbial sources.
Novelty evaluation and profiling
Given the continued rediscovery of known molecules in NP extracts, all the NP screening strategies have been accompanied by implementing efficient, early LC-MS dereplication platforms to identify known compounds in NP DBs containing their spectra (see below). LC-MS has become a preferred method for profiling crude NP extracts and derived fractions for the following reasons: (a) minimal sample preparation, (b) speed, (c) robustness, and (d) high information content [28, 30]. As stated above, this procedure was the first to be called dereplication (or chemical dereplication) and still defines the word in the narrow sense. In spite of technological improvements, activity-guided fractionation is still the most widely used, successful and well-established platform to isolate and characterize active constituents present in microbial or plant extracts sorted by HTS [40-42]. The novelty evaluation process downstream of biological activity-guided HTS typically involves three steps: (i) bioautography, chromatographic separation achieved by HPLC combined with UV and MS detection (LC-DAD-MS) and activity assay; (ii) extended MS analysis including MS-MS spectra, atmospheric pressure chemical ionization (APCI), both in negative and positive mode, and possibly HRMS to calculate elementary composition; and (iii) DB searches using data on biological activity, producer strain, biophysical characterization, and molecular composition. An alternative is to prepare LC-fraction libraries prior to HTS screening to facilitate chemical dereplication and then submit them to screening, as described by Wagenaar [42].
(i) Bioautography
To identify an active component, a NP extract sorted by HTS needs to be bioautographed. The biological activity detected by screening assays needs to be attributed to a chromatographic peak and associated with the UV and MS data generated for this peak by LC-DAD-MS: this is usually done by connecting the HPLC to a fraction collector and testing the activity of each fraction by using the screening assays. Several slightly different systems and processes have been extensively described [43-47]. These require at least one chromatographic step and large amounts of starting biological material. The development of micro-fractionation approaches based on advanced HPLC techniques [18,20,47] has provided the possibility to accelerate the overall process of bioassay-guided fractionation by enabling the systematic separation of complex mixtures using widely applicable protocols. It is important that the bioactivity screening system in use is specific, robust, and sensitive [21, 43]. The system we are currently using is illustrated in Figure 2: a fraction collector is connected to an LC-DAD-MS system (LTQ-Xl- Accela-PDA Surveyor Thermofisher) and, at occurrence, to an evaporative light scattering detector (ESLD); the fractions are collected in 96-well micro-titer plates and subjected to various assays in HTS to reveal which peak corresponds to the active component. Presently, the C18 or C8 reverse-phase columns are the most commonly used HPLC columns for analyzing unknown microbial extracts and profiling metabolites [20,23,28]. They largely apply an elution gradient using a water and methanol/acetonitrile solvent system. Other columns such as HILIC, CN, Amino, C4, Phenyl-Hexil, and chiral matrix are employed for more dedicated use. For example, hydrophilic interaction chromatography (HILIC) provides a solution to fractionate aminoglycoside or very polar compounds [20,28,48]. A splitter and an optimized splitting ratio are required to connect the fraction collector to MS or ELSD detectors. Generic reverse-phase gradients are generally preferred for profiling because they are fully compatible with MS detection using atmospheric pressure ionization sources (ESI or APCI)[28].
Although the biological assays might be very sensitive, a sufficient amount of the fraction corresponding to the active peak still needs to be collected. Separation programs employing LC columns with a diameter of 4.6 mm and 250 mm in length at a flow rate 1 ml/min represent the most common choice. As the ultimate impact is determined by the bioassays, columns of larger diameter (10 mm) are recommended when the fractions need to be directly evaluated in “in vivo” tests, as in the case of the screening performed using the zebrafish animal model [49], or ultra-performance liquid chromatography (UPLC) micro-fractionation in 1536-well plates to screen for inhibitors of protein kinase A activity [48]. Solvent needs to be removed by a centrifugal evaporator before testing the fractions for bioactivity; this step is usually time consuming, the HPLC reverse phases are water rich, and unstable compounds can be degraded easily. The use of a trap column (solid-phase extraction cartridge, SPE) can replace the evaporation process, making it possible to concentrate the unstable molecules [50]. Some substances cannot be captured as a peak by reverse-phase LC chromatography; thus, normal-phase chromatography and direct analysis in real-time MS (Dart-MS) can be used to detect the MS spectrum of compounds directly by preparative thin-layer chromatography (TLC) [51]. A combination of SPE cartridges (LH20,SAX, Oasis MAX, SCX, etc.) can be used in such cases to fractionate and analyze the elution pattern of activity directly [52] or may be combined with sensitive (susceptible)-resistant pair mutant screening [53]. Miniaturized high-throughput SPE and high resolution Fourier transform liquid chromatography MS (HRFT LC-MS) can be employed for discovering new compounds [52].
(ii) Extended MS analysis
LC-MS instruments can be equipped with different ion sources such as ESI, APCI, and atmospheric pressure photo ionization (APPI). The ion source produces ions either by electron ejection, electron capture, cationization, deprotonation, or transfer of a charged molecule from the condensed to the gas phase. In comparison to other ionization sources such as APCI, electronic impact (EI), fast atom bombardment (FAB), and chemical ionization (CI), the techniques of matrix-assisted laser desorption/ionization (MALDI) and ESI have greatly extended MS analyses to a wide range of compounds with an improved sensitivity ranging from the picomole to the zeptomole level. For microbial NP discovery, ESI is the most widely used and a large number of reports using ESI have been published since the 1990s [20]. By employing an ESI ion source, in fact, both positive (PI) and negative (NI) polarities can be analyzed simultaneously and the combination of this information may help to unambiguously show the molecular ions [30]. In addition, ESI can generally detect completely what is inside the sample. The mass analyzer is a critical component for the performance of any mass spectrometer. As MS instruments, the quadrupole-type system, ion trap, time of flight (TOF), and Orbitrap can be used for dereplication. Many studies [13,18,20] now incorporate the use of MS analyzers suitable for accurate mass MSn detection. Instruments such as Orbitrap or TOF have a high mass accuracy, high resolving power, fast scanning rates, and a wide dynamic range. They promote both the discovery of novel compounds that were previously missed by traditional techniques and the detection of already characterized compounds prior to isolation efforts. Finally, HRMS is a very powerful technique; although the instrument is still quite expensive, its direct coupling to LC-MS is increasing among groups performing NP screening [13,24,27,43]. The tool selected for analysis dramatically affects the subset of natural product space that can be visualized (e.g., volatility, polarity, size) and what information can be provided on specific structural features.
(iii) DB searching
Once the MS data on the active fraction are obtained, they are used to identify known compounds in NP DBs that collect MS spectra from known compounds. Today, several commercial DBs are available to implement the dereplication process, the most comprehensive ones being: Chemical Abstracts Natural Products DBs such as Antibase (http://www.user.gwdg.de/Bhlaatsc/antibase.htm), Chapman & Hall Dictionary of Natural Products (CHDNP) (http://dnp.chemnetbase.com), MarinLit (http://www.chem.canterbury.ac.nz/marinlit/marinlit.shtml) and SciFinder (http://scifinder.cas.org/). There are also some freely available DBs such as NIST (http://webbook.nist.gov), METlin (http:// metlin.scripps.edu), and ChemSpider (http://chemspider.com).
To our knowledge, the best solution is to use all or the majority of them, in combination with proprietary DBs (if available) which were often developed through the years by groups involved in NP screening. For instance, we use a proprietary database called ABL [6-7,54], which has been collecting information on bioactive microbial products in the literature and from patents since 1950; ABL contains information on 36000 metabolites derived from actinomycetes and fungi. According to the CHDNP over 246000 natural compounds have been described and approximately 4000 new ones are added each year. The number of metabolites listed in Antibase 2014, for example, is 42950; the Antibase DB is updated yearly, and 800 metabolites were recently added to the Antibase 2013 version. Approximately 24000 marine compounds isolated from 6000 species are listed in MarinLit. Querying for an unknown structure in more than one database reduces the risk of error. It has been estimated that in almost 3% of the structures contained in a database, for example, in Antibase, the elemental compositions are incorrect and that 5% of the structures published annually are not indexed [55]. Generally, it is difficult to identify compounds in the DBs only using MS data and, thus, referring to the UV spectra in addition to MS data can be helpful. The presence of a characteristic UV chromophore can suggest that the unknown compound belongs to a known chemical class. Nowadays, the HPLC or UPLC instruments equipped with a DAD are commercially available with their own UV spectra DB. Unfortunately, though, most of the instruments with their own UV DB cannot be combined with the MS system. If one wants to use an MS instrument and a UV spectrum DB, two PCs are needed for acquisition. Moreover, the UV λmax wavelength records are not available for all the microbial molecules described in the literature. This is one of the reasons why some research groups have made an effort to construct their own DB by accumulating in-house data on LC-UV-MS [18,30,55-58]. In a very useful recent work by the group of Nielsen et al., fungal metabolites were listed with their MS, UV, and retention time (RT) data [17-19, 57]. RT is another parameter not commonly contained in public databases and could be very useful. Boswell et al [59] reported that MS data and RT are orthogonal; therefore, compounds can be identified more efficiently from a combination of RT and LC-MS than only with LC-HRMS. In the novelty evaluation process, the MW of the molecules are compared with those of known compounds. As stated previously, molecular mass is assessed by analyzing the molecular ions and ions adducts; for example, molecules with masses >1000 Da, such as lantibiotics, lipopeptides, and peptaibols, produce doubly and triply charged ions that appear in the scan window of m/z 100–1000. Only few molecules, such as the special cyclic peptides cereulide and valinomycin, which are very strong K+ ionophores, produce only [M+Na]+ and [M+K]+ ion adducts. MS/MS spectra can be easily obtained during micro-fractionation with LC-UV-MS analysis. Ion trap, triple quadrupole, and Q-TOF-MS can acquire MS/MS data, and even single-stage quadrupole and a TOF apparatus can also acquire MS/MS spectra using in-source collision-induced decay. It has been reported that a structure can be elucidated by MS/MS spectra [20,30,60-61], but this is more difficult for NPs with a complex scaffold. Unfortunately, no public, exhaustive DBs exist for the ESI-MS/MS spectra: the only one available is DB Mass Bank, in which 30000 MS/MS spectra have been collected. This DB can be used in a web environment, but the number of microbial natural compounds covered is still too low to be widely useful [62].
Recently, Dorrestein et al. [63-64] suggested that the future of MS in NP discovery could be creating molecular networks as a powerful complement to current dereplication strategies. Molecular networking is an approach in which MS/MS data are organized according to chemical similarity. MS-based molecular networking relies on the observation that structurally similar molecules share similar MS/MS fragmentation patterns. Successful dereplication with molecular networks requires MS/MS spectra of the NP mixture along with MS/MS spectra of known standards, synthetic compounds, or well-characterized organisms, preferably organized into robust databases. This approach can accommodate different ionization platforms, enabling cross-correlations of MS/MS data from ambient ionization, direct infusion, and LC-based methods. A molecular network is a visual representation of molecular relatedness (chemical similarity) of any given set of compounds, not only dereplicating known molecules from complex mixtures, but also capturing related analogs, which constitutes a challenge for many other dereplication strategies. Using this approach 58 molecules, including analogues, were recently dereplicated from marine and terrestrial microbial samples, indicating that molecular networks can be applied to the process of NP screening [63]. A molecular networking algorithm was previously described as an analysis platform upon which new subroutines and visual applications [34, 37] may be added with relative ease, thus allowing further improvements [63-64] such as creating the database now available called “The global natural products social molecular networking” (http://gnps.ucsd.edu), which promises faster dereplication and unique insights as a result of metabolomics comparison.
Figures and Tables
[Click to enlarge]
Quantification and structure elucidation
Although the active constituents present in NP extracts can be identified more quickly as less time is expended for purifying inactive constituents, the bottleneck in NP screening is still the time required to isolate the bioactive, unknown compounds and determine their structure. In general, estimating the amount of metabolite present in the microbial extract (i) can be useful for a further step of structure elucidation (ii).
(i) Quantification
One useful piece of information that can be gained during fractionation and chemical dereplication is the amount of the unknown metabolite present in the microbial extract [20,28,35]. A microbial extract may contain a large amount of low-potency metabolites or a trace amount of a highly potent metabolite. The ability to quantitatively and accurately determine the metabolite’s presence in the dereplication step would represent an advantage for the subsequent purification procedure but is complicated by the low amount of microbial extract sample used in the novelty evaluation step and by the lack of a reference standard. According to the presence of a characteristic chromophoric group in the active molecules LC-UV methods can detect the compound with relatively high sensitivity, but it is complicated to quantitate the results without a standard sample. When the compound has an end absorption, the amount can be guessed to a certain extent, but if interesting bioactive molecules do not have any UV absorption at all, such as some macrolides, quantitation by LC-UV is impossible. Two HPLC compatible detectors can be useful: the previously mentioned ELSD [65] and the chemiluminescent nitrogen detector (CLND) [66]. The first is more versatile than the second, which can only quantify compounds containing nitrogen with a high accuracy and sensitivity. Furthermore, with CLND it is impossible to use nitrogen-containing eluents such as acetonitrile. The principle of ELSD is that the sample is nebulized in the mobile phase of HPLC and, after excluding the large water droplets, solute is obtained as solid particles by heating; finally, the solid particles of the sample are detected by the intensity of scattered light. There is a straight line correspondence in the Log-Log graph in ELSD between the amount of compound and the response, but it is important to note that even if these detectors can weigh the molecules, signals do not correspond to the number of moles [67]. Three other detector systems similar to ELSD have been developed. The nano quantity analyte detector (NQAD) ensures greater sensitivity because the size of solid particles is enhanced by water vapor [68]. The ELSD and NQAD sensitivity is higher than that of refractive index detectors. The Corona’s charged aerosol detection (CAD) system, which employs a measurement principle similar to ELSD, is thought to have a high sensitivity. Solid particles of the solute are obtained by nebulizing the mobile phase of HPLC; then, these particles are charged with a Corona electrode, and the charged particles are quantified using an electrochemical detector [69]. As in ELSD, the signal corresponds to the analyte weight; the relationship between the response and the amount of substance is a straight line in a Log-Log graph. The response from the Corona CAD depends on the composition of mobile phase, such as on acetonitrile content, but if the composition of the mobile phase is maintained at a constant rate before entering the detector with another gradient pump (the so-called reverse gradient), the response reflects only the analyte weight. A second-generation system, the Corona Ultra detector, has been developed to be used with UHPLC. According to [69], NQAD is the most sensitive, and Corona CAD and Corona Ultra are the best analytical solutions in terms of reproducibility. All detectors are less sensitive with semi-volatile molecules (non-polar and with a MW <270 Da).
Quantification of purified natural products can also be obtained by employing NMR (qNMR); here, concentration is given in molar units [70,71] and the peak area of 1H-NMR signal correlates to the number of protons. Recently, the linearity of the receiver gain of NMR instruments has been improved and it is no longer necessary to use an internal or external standard for quantification [72]. Currently, a pulse sequence function, termed electronic reference to access in vivo concentrations (ERETIC) and included as default on the NMR apparatus, can be used to quantify compounds without standard chemicals. Still, in the process of NP screening, generally active compounds are quantified by Corona CAD/Ultra before purification, and qNMR is useful for quantifying already purified compounds (see below).
(ii) Structure elucidation
The method of choice for structural elucidation is NMR spectroscopy since MS alone is not sufficient. The major problem of NMR is sensitivity. Improvements in NMR instrumentation and technology have been made but the amount of sample required for NMR is still in the μM range while for MS it is in pM range. Classically, NMR needs milligram amounts (from few to hundreds of milligrams) of pure compound, depending on the complexity of NP scaffolds.
The application of NMR as an HPLC detector can thus be regarded as an ideal combination for both separation and structural identification of natural products. The advantage of LC-NMR is that not only full structural and stereochemical information can be obtained (by the use of 2D NMR), but it also will detect any hydrogen-containing compound present in the HPLC eluate in a sufficient amount regardless of its structure [29,31,66]. From a historical point of view, the interest in combining separation methods with 1H-NMR spectroscopy arose as early as the end of the 1970s. Owing to the inherent lack of sensitivity of NMR at that time and the problems related to efficient solvent suppression, however, it took almost two decades before LC-NMR started to be used practically to solve analytical problems [20,28,43]. The efficiency of lead discovery would improve dramatically if the structure could be determined from the low amount of samples achieved during fractionation for dereplication. NMR spectroscopy has an intrinsically low sensitivity but microflow NMR [72] and cryo- and microcryo NMR technologies [73-76] only require microgram amounts of sample to acquire 1H and 13C-spectra. The 1H-NMR is also useful to assess purity; common impurities invisible in LC-DAD-MS such as lipids, characterized by low UV absorption, hydrophobicity, and recalcitrance to ionization, can be seen in 1H-NMR. Once an appropriate 1H-NMR spectrum has been obtained, the structure can be elucidated by combining the NMR with UV and MS data. If the compound has some characteristic signals, it is possible to query a specialized DB by entering the number of NMR signals.
LC-NMR techniques are being used more widely, with the potential to eliminate the need of isolating a pure compound before NMR acquisition. It is possible to perform online NMR acquisition from LC using a special flow cell as reviewed in [28-31,77-81]. In 1H-NMR of a mixture sample, clearly different values of signal integrals make it easy to distinguish the individual substances, but when mixed substances show similar signal intensity with very different molecular weights, measuring by diffusion-ordered NMR spectroscopy (DOSY) spectrum is recommended. DOSY is a method of separating the mixed signals by utilizing the difference in the coefficient of molecular diffusion [80-81]. Problems are encountered when diffusion coefficients are similar and/or spectra overlap heavily. However, new high-performance NMR spectrometers are capable of a magnetic field gradient and have the pulse sequence installed by default. As an example, the use of a slow diffusion matrix (micellar sodium laurylsulfate) allowed the resolution of this problem for flavonoid mixtures [82].
Even when high-quality 1H-NMR spectra are obtained, sometimes it is not possible to elucidate the structure. Thus, the various 2D-NMR spectra such as heteronuclear single quantum coherence (HSQC), 1H-detected heteronuclear multiple bond correlation (HMBC), double quantum filtered–COSY (DQF-COSY), total correlation spectroscopy (TOCSY), nuclear Overhauser effect spectroscopy (NOESY), and rotating frame Overhauser effect spectroscopy (ROESY) are usually performed to obtain a planar structure of the compound. Presently, 600MHz NMR equipped with a Cryo Probe (Cold Probe) can measure two-dimensional NMR of the proton in sample amounts of 10 mg. Nonetheless, structural analysis of compounds constituted by many hetero elements and quaternary carbons can still prove difficult even for relatively low-molecular-weight molecules. Wolfender et al. [20] very recently reviewed carefully the current advantages of the different techniques for profiling the NP extracts and suggested LC–DAD-HRMS-SPE-NMR as the most technologically complete platform, facilitating successful dereplication in terms of time, ease, automation, quality, and identification yield. Ideally, the platform is equipped with microvolume, cryogenic, flow probes, or combinations thereof. However, the success of these approaches is dependent on the quality of the spectra to be processed and the efficacy of the algorithms used.
Finally, it is worth mentioning some recent, interesting developments in determining the molecule structure, including stereochemistry, by X-ray crystallography. This technique requires a certain amount of sample; moreover, compound crystallization is not always achieved. It was reported that unknown compounds present in trace amounts can be absorbed in a crystalline sponge, into the tiny crystal of a porous complex, allowing a successful acquisition of crystallographic data [83]. The guest molecules are oriented regularly in the sponge pores and crystallographic analysis can determine the absorbed guest structure along with the host frameworks. This method was suggested as a new tool for elucidating the structure of the small amounts of molecules obtained by HPLC fractionation, and can be applied for compounds with a molecular weight of 500 or less and with a low molecular polarity; the molecular size of the guest molecules should be smaller than the pores. At the moment, this technique is not widely applicable to NPs, but in the future it may become a powerful method for determining the structure of those in trace amounts.
Conclusions
Constructing an appropriate assay system and screening strategy are the most important aspects for biological activity-guided HTS of NPs. The assay system and the screening strategy include at different phases dereplication steps that are crucial first for widening the upstream biological diversity of the NP libraries and then for speeding up the novelty evaluation process downstream to the bioassay-guided HTS. MS and NMR are the two predominant analytical platforms for detecting and identifying metabolites; they should still be used complementarily, the first being essential for the novelty evaluation, the second indispensable for structure elucidation. The major weaknesses of MS represent the major strengths of NMR spectroscopy, and vice versa. In Table 1 we summarize the specific advantages of the two approaches. We have reported on the recent rapid development of MS and NMR instruments and technology, but a crucial aspect is combining them with advanced chromatographic methods and sampling devices which progressively introduce/consolidate their use in understanding microbial and chemical diversity during the process of NP screening. Complex samples might become directly accessible by applying novel, integrated, analytical approaches, with the aim of discovering novel NPs. Concurrently, the rate of data becoming available is definitively increasing, making it essential to organize the information in well-constructed and exhaustive databases to enable cross talk and assist with novelty evaluation and structure elucidation. This is without doubt the other extremely demanding and evolving need that is currently emerging in the field.
In conclusion, other fields related to NP screening are currently benefiting highly from the analytical approaches described in this mini-review, such as deciphering chemical communications in microbial ecology and profiling metabolomics of living organisms and ecosystems [32-34,36-38,84]. Alternatively, recent progress in our understanding of biosynthesis and regulation of specialized metabolites and the increasing information from the genomics of their producer strains is being used more and more to predict and assist novelty evaluation structure elucidation of novel bioactive NPs [85-86]. Although the latter topics are beyond the scope of this mini-review, we would like to emphasize that the portfolio of analytical methods is becoming more and more an essential and integrated part of the interdisciplinary approach evolving in the postgenomic era for discovering novel drugs from NP sources.
Acknowledgments
We thank FIIRV colleagues.
References
1. Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 (2012). [CrossRef]
2. Berdy J. Bioactive microbial metabolites.2005 J. Antibiot. 58, 1-26 (2005). [CrossRef]
3. Li JWH, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science 325, 161–165 (2009). [CrossRef]
4. Mark S, Butler A, Robertson AB, Cooper M. Natural product and natural product derived drugs in clinical trials. Nat. Prod. Rep. 4, 1612-1661 (2014). [CrossRef]
5. Davies J. Specialized microbial metabolites: functions and origins. J. Antibiot. 66, 361-336 (2013). [CrossRef]
6. Marinelli F. From microbial products to novel drugs that target a multitude of disease indication. Methods in Enzymology 458, 29-58 (2009).
7. Marinelli F, Marcone GL. Small Molecules – Microbial Secondary Metabolites. In: Comprehensive Biotech. Volume 3, Second Edition, Edited by Murray Moo-Young , Elsevier BV, pg. 285–297 (2011).
8. Luo Y, Cobb RE, Zhao H. Recent advances in natural product discovery. Curr. Opin. Biotech. 30, 230-237 (2014). [CrossRef]
9. Drewry DH, Macarron R. Enhancements of screening collections to address areas of unmet medical need: an industry perspective. Curr. Opin. Chem. Biol. 14, 289-298 (2010). [CrossRef]
10. Baltz RH. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 33, 507-513 (2006). [CrossRef]
11. Baltz RH. Natural product discovery and development at Eli Lilly and Company: One scientists view. SIM News 55, 5-16 (2005).
12. Ito T, Masubuchi M. Dereplication of microbial extracts and related analytical technologies. J. Antibiot. 67, 353-360 (2014). [CrossRef]
13. Sidebottom A, Carlson EE. A reinvigorated era of bacterial secondary metabolite. Curr.Opin. Chem. Biol. 24, 104-111 (2014). [CrossRef]
14. Stewart EJ. Growing unculturable bacteria. J. Bacteriol. 194, 4151-4160 (2012). [CrossRef]
15. Langlykke A. CRC Handbook of antibiotic compound (IV), Edited by Bardy J et al. CRC, Boca Raton, FL, (1980).
16. Van Middlesworth F , Carnell RJP. Dereplication and partial identification of natural products. Nat. Prod. Isolation. 4, 279-327 (1998). [CrossRef]
17. Nielsen K. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography–UV–mass spectrometry methodology. J. Chromatogr. A 1002, 111–136 (2003). [CrossRef]
18. Nielsen, KF, Mansson M, Rank C, Frisvad JC, Larsen TO. Dereplication of microbial natural products by LC-DAD-TOFMS. J. Nat. Prod. 74, 2338–2348 (2011). [CrossRef]
19. Klitgaard A, Iversen A, Andersen MR, Larsen TO, Frisvad JC, Nielsen KF. Aggressive dereplication using UHPLC–DAD–QTOF: screening extracts for up to 3000 fungal secondary metabolites. Anal. Bioanal. Chem. 406, 1933–1943 (2014). [CrossRef]
20. Wolfender JL, Marti G, Thomas A, Bertrand S. Current approaches and challenges for the metabolite profiling of complex natural extracts J. Chromatogr A. 1382C, 136-164 (2015). [CrossRef]
21. Fedorenko V, Genilloud O, Horbal L et al. Antibacterial discovery and development: from gene to product and back. BioMed Res. Int. In press (2015).
22. Zhang H, Lee YK, Zhang W, Lee HK. Culturable actinobacteria from the marine sponge Hymeniacidon perleve: isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Antonie Leeuwenhoek 90, 159-169 (2006). [CrossRef]
23. Gonzalez-Sabin J. Natural products: Back to the future in drug discovery. Biochem. Pharmacol. 1, 1-3 (2012). [CrossRef]
24. Larsen TO, Smedsgaard J, Nielsen KF, Hansen ME, Frisvad JC, Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Nat. Prod. Rep. 22, 672-695 (2005). [CrossRef]
25. Lanoot B, Vancanneyt M, Dawyndt P, Cnockaert M, Zhang J, Huang Z, Liu Z, Swings J. BOX-PCR Fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus. Syst. Appl. Microbiol. 27, 84–92 (2004). [CrossRef]
26. Lanoot B, Vancanneyt M, Vandemeulebroecke K et al. Grouping of streptomycetes using 16S-ITS RFLP fingerprinting. Res. Microbiol. 156, 755–762 (2005). [CrossRef]
27. Forner D, Berrue F, Corre H, Duncan K, Kerr RG. Chemical dereplication of marine actinomycetes by liquid chromatography–high resolution mass spectrometry profiling and statistical analysis. Analytica Chimica Acta 805, 70-79 (2013). [CrossRef]
28. Bucar F, Wube A, Schmid M. Natural product isolation – how to get from biological material to pure compounds Nat. Prod. Rep. 30, 525–545 (2013). [CrossRef]
29. Halabaki M, Vougogiannopoulou K, Mikrosn E, Skaltsounis et al. Recent advances and new strate-gies in the NMR based identification of natural products. Curr. Opin. Biotech. 25, 51-59 (2014). [CrossRef]
30. Jarmusch AK , Cooks RG. Emerging capabilities of mass spectrometry for natural products. Nat. Prod. Rep. 31, 730–738 (2014). [CrossRef]
31. Simmler C, Napolitano JG, McAlpine JB, Chen SN, Pauli GF. Universal quantitative NMR analysis of complex natural samples Curr. Opin. Biotech. 25, 1-7 (2014). [CrossRef]
32. Nemes P, Barton AA, Vertes A. Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal. Chem. 81, 6668-6675 (2009). [CrossRef]
33. Watrous J, Roach P, Heath H, Alexandrov T, Laskin J, Dorrestein PC. Metabolic profiling directly from the Petri dish using nanospray desorption electrospray ionization imaging mass spectrometry. Anal. Chem. 85, 10385-10391 (2013). [CrossRef]
34. Watrous J, Roach P, Heath H et al. Mass spectral molecular networking of living microbial colonies Proc. Natl. Acad. Sci. 109, 1743-1752 (2012). [CrossRef]
35. Zachariasova M, Cajka T, Godula M, Malachova A, Veprikova Z, Hajslova J. Analysis of multiple mycotoxins in beer employing (ultra)-high resolution mass spectrometry. Rapid Commun. Mass Spectrom. 24, 3357–3367 (2010). [CrossRef]
36. Vaclavik L, Zachariasova M, Hrbek V, Hajslova J. Analysis of multiple mycotoxins in cereals under ambient conditions using direct analysis in real time (DART) ionization coupled to high resolution mass spectrometry. Talanta 82, 1950–1957 (2010). [CrossRef]
37. Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol. 9, 683–694 (2011). [CrossRef]
38. Hsu C, ElNaggar MS, Peng Y et al. Real-time metabolomics on living microorganisms using ambient electrospray ionization flow-probe. Anal. Chem. 85, 7014–7018 (2013). [CrossRef]
39. Kai M, Gonzalez I, Genilloud O, Singh SB, Svatos A. Direct mass spectrometric screening of antibiotics from bacterial surfaces using liquid extraction surface analysis. Rapid Commun. Mass Spectrom. 26, 2477–2482 (2012). [CrossRef]
40. Lang G, Mitova MI, Ellis G et al. Bioactivity profiling using HPLC/microtiter-plate analysis: application to a New Zealand marine alga-derived fungus, Gliocladium sp. J. Nat. Prod. 69, 621–624 (2006). [CrossRef]
41. Johnson TA, Sohn J, Inman WD et al. Natural product libraries to accelerate the high-throughput discovery of therapeutic leads. J. Nat. Prod. 74, 2545–2555 (2011). [CrossRef]
42. Wagenaar MM. Pre-fractionated microbial samples – the second generation natural products library at Wyeth. Molecules 13, 1406–1426 (2008). [CrossRef]
43. Potterat O, Hamburger M. Concepts and technologies for tracking bioactive compounds in natural product extracts: generation of libraries, and hyphenation of analytical processes with bioassays. Nat. Prod. Rep. 30, 546–564 (2013). [CrossRef]
44. Michel T, Halabalaki M, Skaltsounis AL. New concepts, experimental approaches, and dereplication strategies for the discovery of novel phytoestrogens from natural sources. Planta Med. 79, 514–532 (2013). [CrossRef]
45. Marston A. Role of advances in chromatographic techniques in phytochemistry. Phytochem. 68, 2786–2798 (2007). [CrossRef]
46. Curtis JH, Beutler JA. Matching the power of high throughput screening to the chemical diversity of natural products, Nat. Prod. Rep. 30, 1284-1298 (2013). [CrossRef]
47. Giera M, Heus F, Janssen L, Kool J, Lingeman H, Irth H. Microfractionation revisited: A 1536 well high resolution screening assay. Anal Chem 81, 5460–5466 (2009). [CrossRef]
48. Kumar P, Rubies A, Companyó R, Centrich F. Hydrophilic interaction chromatography for the analysis of aminoglycosides. J Sep Sci 35, 498-504 (2012). [CrossRef]
49. Bohni N, Cordero-Maldonado ML, Maes J et al. Integration of microfractionation, qNMR and zebrafish screening for the in vivo bioassay-guided isolation and quantitative bioactivity analysis of natural products. PLoS One 8, 5, e64006 (2013). [CrossRef]
50. Johansen KT, Wubshet SG, Nyberg NT. HPLC-NMR revisited: using time-slice high-performance liquid chromatography-solid-phase extraction-nuclear magnetic resonance with database-assisted dereplication. Anal. Chem. 85, 3183–3189 (2013). [CrossRef]
51. Cheng SC, Huang MZ, Shiea J. Thin layer chromatography/mass spectrometry. J. Chromatogr. A. 1218, 2700-2711 (2011). [CrossRef]
52. Mansson M, Phipps RK, Gram L, Munro MH, Larsen TO, Nielsen KF. Explorative solid-phase extraction (E-SPE) for accelerated microbial natural product discovery, dereplication, and purification. J. Nat. Prod. 73,1126–1132 (2010). [CrossRef]
53. Singh SB, Zhang C, Zink DL et al. Occurrence, distribution, dereplication and efficient discovery of thiazolyl peptides by sensitive-resistant pair screening. J. Antibiot. 66, 599–607 (2013). [CrossRef]
54. Lazzarini A, Cavaletti L, Toppo G, Marinelli F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie van Leeuwenhoek, 78, 3-4, 399-405 (2001). [CrossRef]
55. Lopez-Perez JL, Theron R, del Olmo E, Dıaz D. NAPROC-13: a database for the dereplication of natural product mixtures in bioassay-guided protocols. Bioinformatics 23, 3256–3257 (2007). [CrossRef]
56. Holt TG. The design and development of an integrated natural products screening database. J. Biomol. Screen. 5, 421–433 (2000). [CrossRef]
57. Klitgaard A, Iversen A, Andersen MR, Larsen TO, Frisvad JC, Nielsen KF. Aggressive dereplication using UHPLC–DAD–QTOF: screening extracts for up to 3000 fungal secondary metabolites. Anal. Bioanal. Chem. 406, 1933–1943 (2014). [CrossRef]
58. Kato N, Takahashi S, Nogawa T, Saito T., Osada H. Construction of a microbial natural product library for chemical biology studies. Curr. Opin. Chem. Biol. 16,101–108 (2012). [CrossRef]
59. Boswell PG, Schellenberg JR, Carr PW, Cohen JD, Hegeman AD. A study on retention ‘projection’ as a supplementary means for compound identification by liquid chromatography-mass spectrometry capable of predicting retention with different gradients, flow rates, and instruments. J. Chromatogr. A 1218, 6732–6741 (2011). [CrossRef]
60. Ridder L., van der Hooft JJ, Verhoeven S, de Vos RC, Bino RJ, Vervoort J. Automatic chemical structure annotation of an LC-MS(n) based metabolic profile from green tea. Anal. Chem. 85,6033–6040 (2013). [CrossRef]
61. Konishi Y, Kiyota T, Draghici C et al. 2007 Molecular formula analysis by an MS/MS/MS technique to expedite dereplication of natural products. Anal. Chem. 79, 1187–1197 (2007). [CrossRef]
62. Horai H, Arita M, Kanaya S et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714 (2010). [CrossRef]
63. Yang JY, Sanchez LM, Rath CM et al. Molecular networking as a dereplication strategy J. Nat. Prod. 76, 1686–1699 (2013). [CrossRef]
64. Winnikoff JR, Glukhov E, Watrous J, Dorrestein PC, Gerwick WH. Quantitative molecular networking to profile marine cyanobacterial metabolomes J. Antibiot. 67, 105–112 (2014). [CrossRef]
65. Cremin PA, Zeng L. High-throughput analysis of natural product compound libraries by parallel LC-MS evaporative light scattering detection. Anal. Chem. 74,5492–5500 (2002). [CrossRef]
66. Lane S, Boughtflower B, Mutton I et al. Toward single-calibrant quantification in HPLC. A comparison of three detection strategies: evaporative light scattering, chemiluminescent nitrogen, and proton NMR. Anal. Chem. 77, 4354–4365 (2005).[CrossRef]
67. Adnani N, Michel CR, Bugni TS. Universal quantification of structurally diverse natural products using an evaporative light scattering detector. J. Nat. Prod. 75, 802–806 (2012). [CrossRef]
68. Hutchinson JP, Li J, Farrell W, Groeber E, Szucs R, Dicinoski G, Haddad PR. Comparison of the response of four aerosol detectors used with ultra high pressure liquid chromatography. J. Chromatogr. A 1218, 1646–1655 (2011). [CrossRef]
69. Vervoort N, Daemen D, Torok G. Performance evaluation of evaporative light scattering detection and charged aerosol detection in reversed phase liquid chromatography. J. Chromatogr. A 1189, 92–100 (2008). [CrossRef]
70. Hasada K, Yoshida T, Yamazaki T, Sugimoto N, Nishimura T, Nagatsu A, Mizukami H. Quantitative determination of atractylon in Atractylodis rhizoma and Atractylodis lanceae rhizoma by 1H-NMR spectroscopy. J. Nat. Med. 64, 161–166 (2010). [CrossRef]
71. Pauli GF, Jaki BU, Lankin DC. A routine experimental protocol for qHNMR illustrated with taxol. J. Nat. Prod. 70, 589–595 (2007). [CrossRef]
72. Farrant, RD, Hollerton JC, Lynn SM, Provera S, Sidebottom PJ, Upton RJ. NMR quantification using an artificial signal. Magn. Reson. Chem. 48, 753–762 (2010). [CrossRef]
73. Eldridge GR, Vervoort HC, Lee CM et al. High-throughput method for the production and analysis of large natural product libraries for drug discovery. Anal. Chem. 74, 3963–3971 (2002). [CrossRef]
74. Williams RB, Martin SM, Hu JF et al. Isolation of apoptosis-inducing stilbenoids from four members of the Orchidaceae family. Planta Med 78, 160–165 (2012). [CrossRef]
75. Johansen KT, Wubshet SG, Nyberg NT, Jaroszewski JW. From retrospective assessment to prospective decisions in natural product isolation: HPLC-SPE-NMR analysis of Carthamus oxyacantha. J. Nat. Prod. 74, 2454–2461 (2011). [CrossRef]
76. Dalisay DS, Rogers EW, Edison AS, Molinski TF. Structure elucidation at the nanomole scale. 1. Trisoxazole macrolides and thiazole-containing cyclic peptides from the nudibranch Hexabranchus sanguineus. J. Nat. Prod. 72, 732–738 (2009). [CrossRef]
77. Jaroszewski JW. Hyphenated NMR methods in natural products research, Part 1:direct hyphenation. Planta Med. 71, 691–700 (2005). [CrossRef]
78. Jaroszewski JW. Hyphenated NMR methods in natural products research, Part 2: HPLC-SPE-NMR and other new trends in NMR hyphenation. Planta Med. 71,795–802 (2005). [CrossRef]
79. Williamson RT, Chapin EL, Carr AW et al. New diffusion-edited NMR experiments to expedite the dereplication of known compounds from natural product mixtures. Org. Lett. 2, 289–292 (2000). [CrossRef]
80. Lang G, Mayhudin NA, Mitova MI et al. Evolving trends in the dereplication of natural product extracts: new methodology for rapid, small-scale investigation of natural product extracts.J. Nat. Prod. 71, 1595-1599 (2008). [CrossRef]
81. Johansen KT, Wubshet SG, Nyberg NT. HPLC-NMR revisited: using time-slice high-performance liquid chromatography-solid-phase extraction-nuclear magnetic resonance with database-assisted dereplication. Anal. Chem. 85, 3183–3189 (2013). [CrossRef]
82. Cassani J, Nilsson M, Morris GA. Flavonoid mixture analysis by matrix-assisted diffusion-ordered spectroscopy. J. Nat. Prod. 75, 131–134 (2012). [CrossRef]
83. Inokuma Y, Yoshioka S, Ariyoshi Y et al. X-ray analysis on the nanogram to microgram scale using porous complexes. Nature 495, 461–466 (2013). [CrossRef]
84. Pettit RK. Mixed fermentation for natural product drug discovery. Appl. Microbiol. Biotechnol. 83, 19-25 (2009). [CrossRef]
85. Thaker MN, Wang W, Spanogiannopoulos P, Waglechner N, King AM, Medina R, Wright GD. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 31, 922-927 (2013). [CrossRef]
86. Challis GL. Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 41, 219-232 (2014). [CrossRef]
All site content, except where otherwise noted, is licensed under a Creative Commons Attribution 4.0 License