OPEN-ACCESS PEER-REVIEWED
REVIEW
Mohammad Sharif Khan* and Jannatul Azmir
Thayer School of Engineering, Dartmouth College, Hanover, NH, 03755, USA.
Journal of Applied Bioanalysis. Vol.6. No.3. pages 97-106 (2020).
Published 15 August 2020. https://doi.org/10.17145/jab.20.012 | (ISSN 2405-710X).
*Correspondence: Khan MS Thayer School of Engineering, Dartmouth College, 14 Engineering Drive, Hanover, NH, 03755, USA. Phone: +1 603 646 2230.
Citation:
Khan MS and Azmir J. Multi-omics for Biomedical Applications. J Appl Bioanal 6(3), 97-101 (2020).
Open-access and Copyright:
©2020 Khan MS and Azmir J. This article is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Funding/Manuscript writing assistance:
The authors have no financial support or funding to report and they also declare that no writing assistance was utilized in the production of this article.
Competing interest:
The authors have declared that no competing interest exist.
Article history:
Received: 21 February 2020, Revised 19 April 2020, Accepted 22 April 2020.
Abstract
Multi-omics is a rising filed in omics science. Despite progress in the development of an appropriate single omics, in-particular for biomedical problem solving; the holistic look at the complex nature of the human cell, disease, and other biochemical pathways remain undiscovered. The multi-omics platform considered the most integrated system currently available to obtain and measure the biochemical data-driven information for biomedical problems. The current review will look at the factors that play important roles in the rise of the multi-omics fields and its application in biomedical studies.
Introduction
Sometimes a few similar good things, when combined, can create a great outcome! The integrated omics or multi-omics has done this by opening up a holistic look at the biological system by combining multiple-omics systems [1]. The “Omes” table at Yale University records the first study with the “omics” keyword published in 1938 in Pubmed [2].
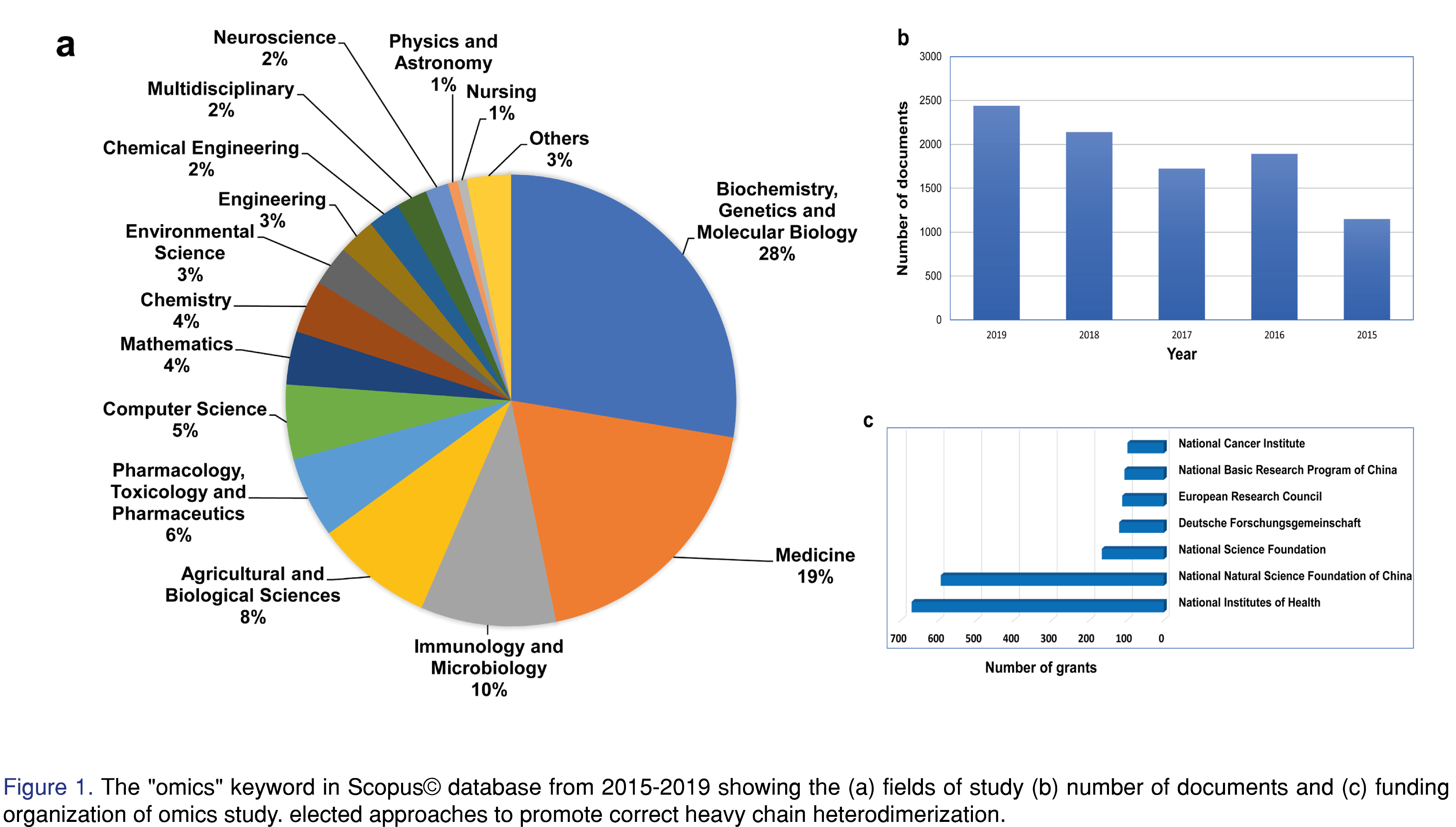
The expansion of the field was so rocketed and within a short period of time, the field now has a large number of branches includes genomics, proteomics, transcriptomics, metabolomics, and so on. The non-biological expansion of omics fields are also wide e.g. foodomics, volatolomics, phytochemomics, and many more. A search in the Scopus database using “omics” keyword during 2015-2019, has illustrated the trend of omics, based on 9342 documents-clearly showing the rise of this field. The main field of studies was dedicated to the biochemical and biomedical sciences whereas these studies were funded by the major world research organizations. During the last five years, almost a 40% increase in omics work was recorded. Among those, 20% of the studies were concentrated on the multi-omics approach, which is an indication of the expanding multi-omics field (Figure 1).
In this mini-review, we will look at the important factors of the multi-omics field and then we’ll discuss the applications of multi-omics for solving of biomedical problems.
Driving forces of multi-omics fields
One of the primary reasons for the expanding multi-omics fields during the last 10 years was the development of high-throughput and high-resolution technology for biological problem-solving [3]. One of the examples is expression array, a method to identify candidate marker genes of disease could generate an overwhelming amount of data of hundreds of thousands of genes in a single run [4]. A second important innovation was the development of high-resolution mass spectroscopy. The HR-MS has emerged with extensive identification power that has become an essential part of environmental, biochemical, and chemical sciences [5]. Figure 2 presents a schematic to show the application of the high-throughput and high-resolution methods in omics fields.
The development of data science is the most critical and meaningful boost to the multi-omics field. The integration of the huge amount of data into a meaningful conclusion was not successful until the development of machine learning, artificial intelligence, and data science [6]. The author refers to multiple well-written reviews for more computer solutions serving the multi-omics field [6-11]. A few main challenges of computer solutions remain; limited biological knowledge guided integration and the heterogeneity of the data from different omics sources.
Application of Multi-omics in biomedical research
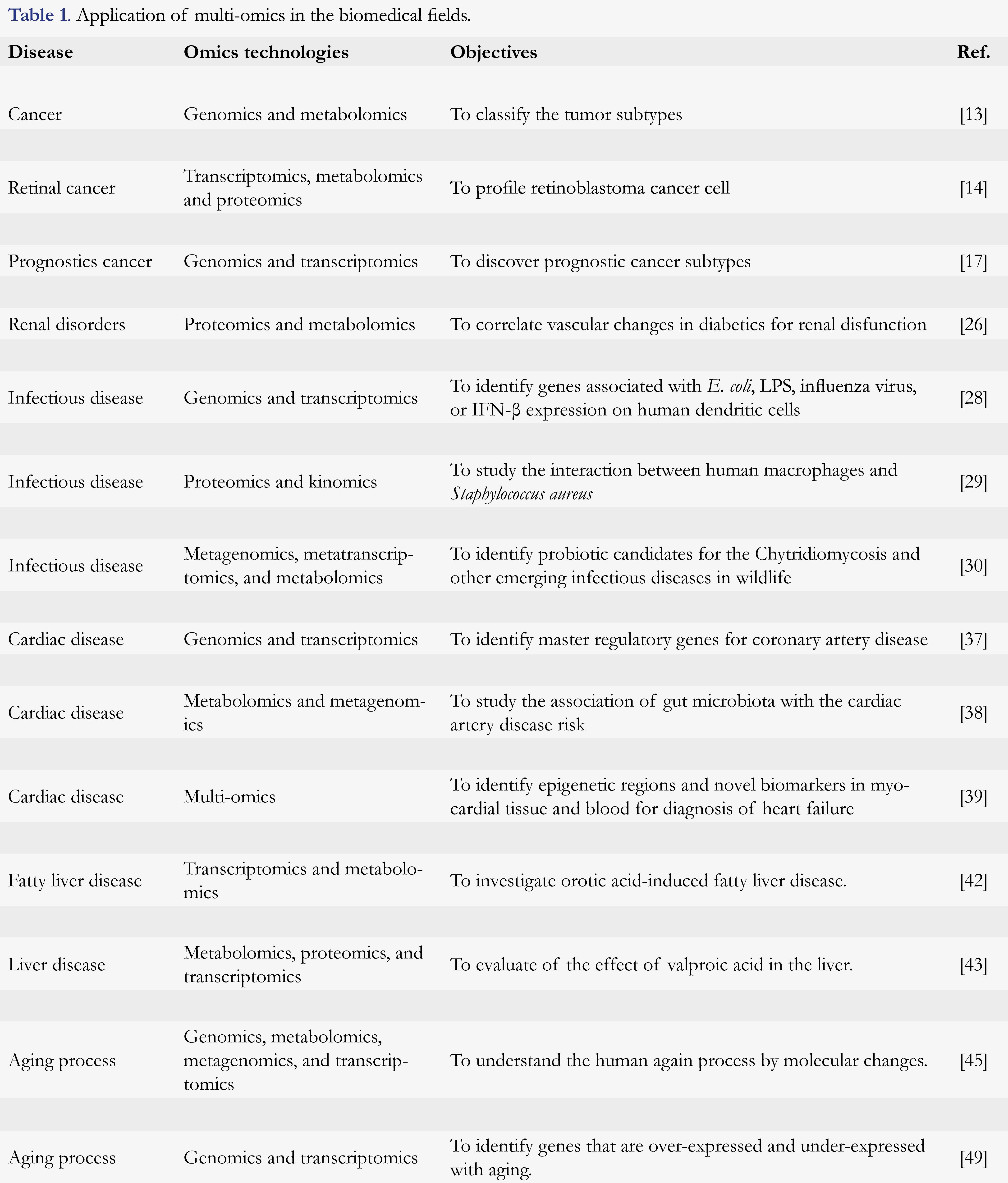
Cancer research
Chronic kidney disease
Infectious disease
Host-pathogen interaction in infectious disease is an important aspect for the understanding disease biomarkers, prognosis, and treatment [27]. Multi-omics studies were used in this field particularly tuberculosis (TB), autoimmune disease, and HIV. Genomics and transcriptomics were used to stimulate with Escherichia coli (E. coli) LPS, influenza virus, or IFN-ß expression on human dendritic cells. A set of 121 genes were found associated with those exposures in that study [28]. The human macrophages and Staphylococcus aureus interactions in terms of proteomics and kinomics were studied. Authors have reported the major macrophage signaling pathways that are triggered by pathogens [29]. An interesting study used metagenomics, metatranscriptomics, or metabolomics application of multi-omics on the identification of probiotic candidates for the Chytridiomycosis and other emerging infectious diseases in wildlife [30].
A longitudinal multi-omics study was conducted on more than 100 healthy and pre-diabetics volunteers for 4 years to track the changes in transcriptomes, metabolomes, cytokines, and proteomes, as well as changes in the microbiome [31]. The result has broadened the understanding of the association between prediabetes biological stages and the identification of inflammation markers in immune signaling. A number of large scale association studies based on an omics platform to understand the pathophysiology of HIV have also been reported [32]. These studies help to understand the link between the complex HIV infection process and the interpretation of actionable therapeutic or diagnostic targets.
Cardiac disease
Cardiovascular diseases are the leading causes of morbidity and mortality worldwide [33]. Coronary heart disease has found associated with a group of 150 genomes in a large scale genomics study [34]. The expression of those genetic loci and the integration of multi-omics data transcriptome, epigenome, proteome, metabolome were reported in the literature [35]. A multi-omics study integrating transcriptomics, proteomics, and proteome study was conducted to evaluate their synergistic effect on the in-vitro cardiac hypertrophy model in mice [36]. That study identified 70 candidate disease signatures in the steady-state transcript and protein abundance.
A genomic and transcriptomic study identified 3 master regulatory genes for Coronary Artery Disease (CAD) patients in a population of Europe; Stockholm Atherosclerosis Gene Expression (STAGE) study [37]. Another study by Feng et al. utilized metabolomics and metagenomics to study the association of gut microbiota with the CAD risk. Another study found that GlcNAc-6-P, mannitol, and 15 plasma cholines were associated with a higher risk of CAD and showed a correlation of the Clostridium sp. and Streptococcus sp. in the intestine [38]. Diagnosis for myocardial dysfunction and heart failure is an important indicator for care and treatment control. A multi-omics study in myocardial tissue and blood has identified epigenetic regions and novel biomarkers for diagnosis of heart failure. The result has reported a set of 517 epigenetic loci and CpGs makers of the heart failure diagnosis [39]. A multi-omics study on CAD not only helps to understand the mechanism of genetic connection but also helps to identify the key drivers and pathways that contribute to the risk [35]. For example, an integrated network has been developed using genome-wide association studies and identified a set of 30 related to CAD [40].
Others
The rise of the omics technologies over the last few years has lead to a better understanding of the mechanism of diet in metabolic regulation and overall health [41]. As a result, several papers integrating the multiple omics in nutritional research has been reported. Orotic acid-induced fatty liver disease was investigated using transcriptional and metabolic levels [42]. The study finding indicated a few metabolic pathways that demonstrate the association of orotic acid with fatty liver disease. Another example of the multi-omics study by metabolomics, proteomics, and transcriptomics is the evaluation of the effect of valproic acid in the liver. The result indicated a perturbation in glycogenolysis pathway by two proteins, glycogen phosphorylase and amylo-1,6-glucosidase different from the control [43]. Another nutritional multi-omics study explores the relation of arachidonate-enriched diet over the eicosapentaenoic (EPA)/docosahexaenoic (DHA) diet, to find any differential response on hepatic lipid metabolism [44].
Molecular changes during the aging process have an important significance in the preservation and treatment of the aging person. The multi-omics study that links genomics, metabolomics, metagenomics, and transcriptomics was used to understand the human again process [45]. The study showed the importance of the relationship between epigenetic factors such as histone modification and DNA methylation in aging [46]. A lipidomics study has suggested a strong association with lipid profile with the longevity [47]. On the other hand, an in-vitro study with rat brain has illustrated the dynamics of changes in the dark matter of the genome [48]. A combined genomic and transcriptomic study has revealed a set of 56 genes that are overexpressed with aging and 17 genes that are under-expressed with age [49].
Figures and Tables
[Click to enlarge]
Tools and methods used in the Multi-omics
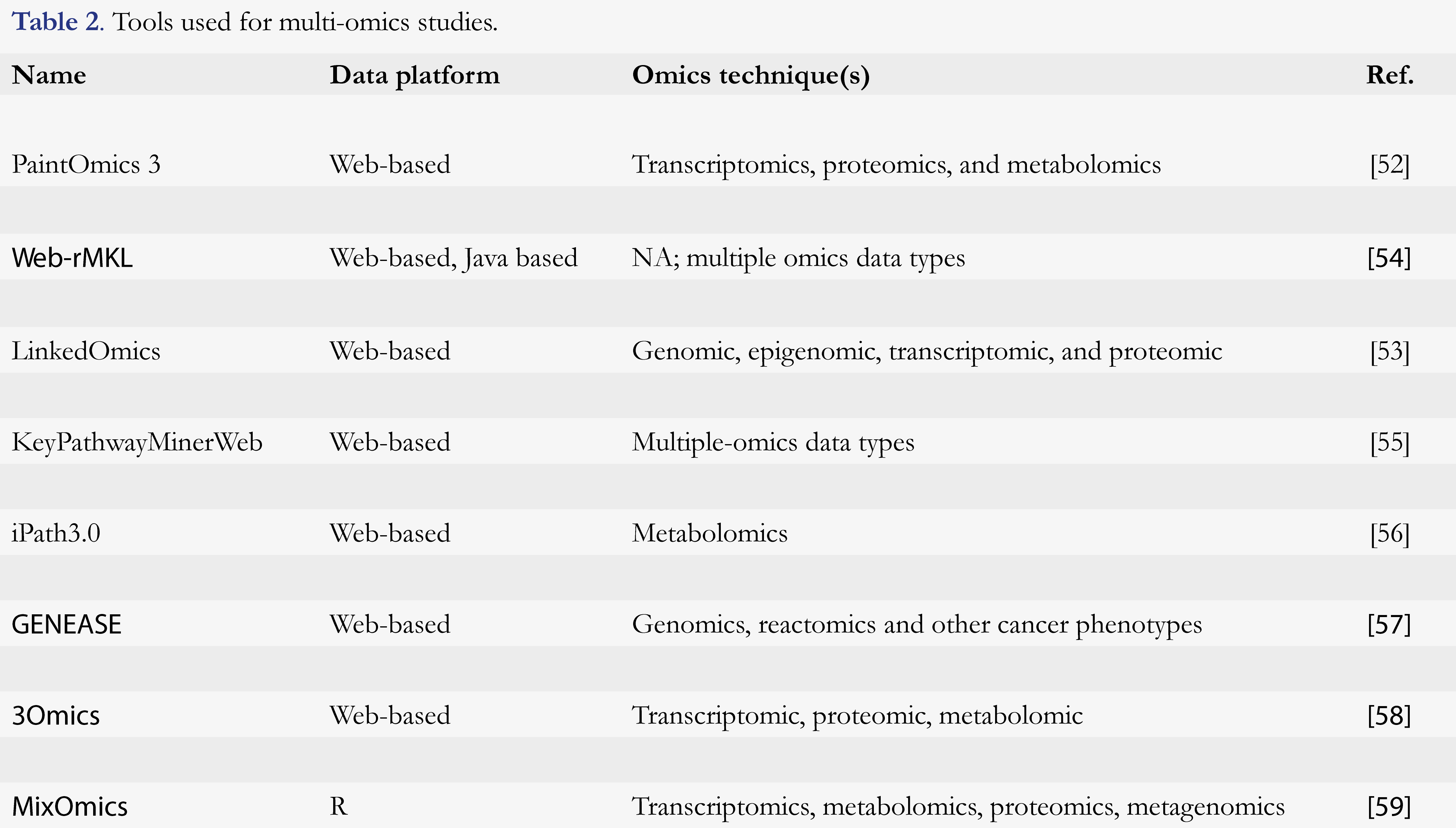
Understanding complex biological problems remains a quest for many omics research. With technological advances, we could now have a huge amount of data from different levels of probes for a better and comprehensive look of a disease. Although the integration of these heterogeneous data remains a major challenge in multi-omics fields, an immense effort by the data scientist and developer has been given. There are a number of different data integration methods and tools have been developed and applied to the multi-omics dataset (Table 2). These tools are able to integrate DNA sequence, RNA expression, methylation patterns, genome, transcriptome, and proteome, and many more [50, 51].
The web-based method for multi-omics data integration and visualization is a recent and modern approach to tackle data integration tools. This process allows users to create a job and submit the data to understand the interconnection between the multi-layer omics dataset. PaintOmics 3 could be an example [52]. This web tool uses the KEGG database to show the integration of the biological pathway. Another good example is the LinkedOmics, which have a billion data points combining mass spectrometry (MS)-based global proteomic and cancer genomic datasets. Users can explore the link between these datasets and generate hypotheses for experimental validation [53]. Table 2 lists a few other examples of these types of tools developed for multi-omics integration.
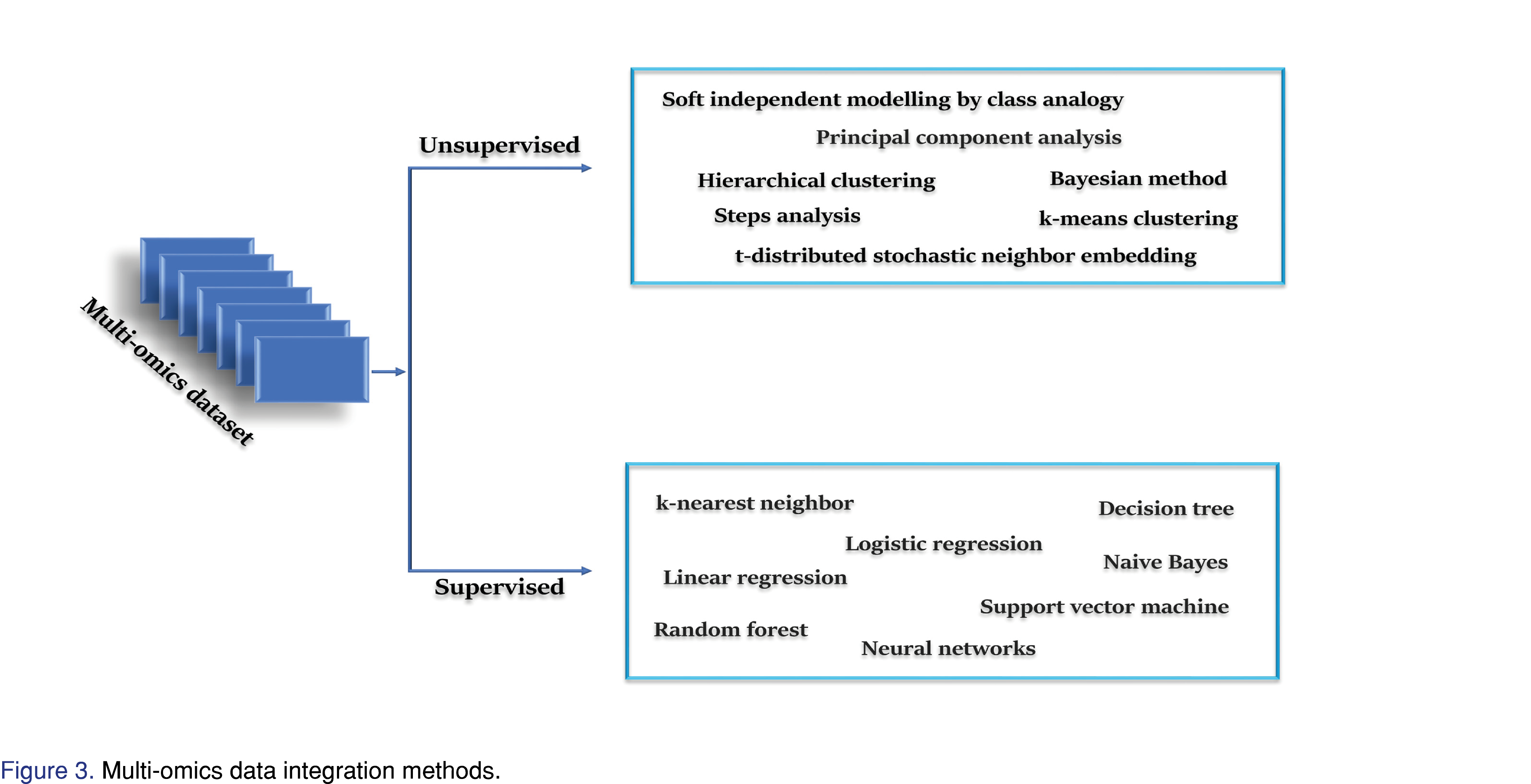
The mathematical integration of the different sources of data with different noise levels, different variables, non–comparable data types, and many other data issues are also challenging in the multi-omics studies. A number of methods have been developed in this regard and mainly divided into two methods; 1) unsupervised data integrating, a method that does not assign the response variables before analysis and 2) supervised data integration, which assigns the response variables before the analysis [6] (Figure 3).
Machine learning, Artificial intelligence, Bayesian method, Network-based analysis, Kernel method, and many more mathematical methods and their mathematical background have been reported in the literature [6,7]. The second consideration is the platform or software used for the data analysis. Some of these tools are based on web-servers, some on C++, R, Matlab, Python, and many more. This field of multi-omics method and tools are in the developmental stage and many new methods are appearing in the literature.
Future Perspectives
A growing number of multi-omics studies have been reported in the last couple of years, incorporating many different omics platforms, new technology, tools, and methods for data integration. The multi-omics process provided unprecedented utility and benefits that could not be provided by any single omics approach. This is now considered the most comprehensive and holistic approaches for analyzing the samples and understanding between multiples biological phenomena by visualization, linking multiple biological pathways, finding an association between genotypes and phenotypes, and many more. The omics platform has opened a new era in biomedical science and the combination of the multi-omics is considering the future in this field.
As the multi-omics platform is getting more mature in terms of the tools and resources, it will facilitate the development of newer fields. Multi-omics will help to understand the link between the molecular and clinical characteristics of the disease, the discovery of biomarkers by pathway analysis, and observed the association from a large scale mechanistic study. The ongoing effort on the multi-omics and leveraging integrative strategies would further improve the application of this filed on the biomedical problems solving.
Conclusions
This mini-review provides a concise overview of the multi-omics platform. The motivation of the multi-omics, its application in biomedical problem solving has been discussed here. The omics platform integration tools and the methods that require special data science background have also been illustrated. The multi-omics is so far the most holistic analysis tool and would be the future of the omics fields.
References
- Vilanova C. and Porcar M. Are multi-omics enough? Nat Microbiol 1(8), 1-2. (2016).
https://doi.org/10.1038/nmicrobiol.2016.101 - Yale University, Gerstein Lab. OMES Table. Available from: http://bioinfo.mbb.yale.edu/what-is-it/omes/omes.html.
- Zoldoš V, Horvat T, Lauc G. Glycomics meets genomics, epigenomics and other high throughput omics for system biology studies. Curr Opin Chem Biol17(1), 34-40 (2013).
https://doi.org/10.1016/j.cbpa.2012.12.007 - Schulze A and Downward J. Navigating gene expression using microarrays-a technology review. Nat Cell Biol 3(8), E190 (2001).
https://doi.org/10.1038/35087138 - Marshall AG and Hendrickson CL. High-resolution mass spectrometers. Annu Rev Anal Chem 1, 579-599 (2008).
https://doi.org/10.1146/annurev.anchem.1.031207.112945 - Huang S, Chaudhary K, Garmire LX. More Is Better: Recent Progress in Multi-Omics Data Integration Methods. Front Genet 8(84) (2017).
https://doi.org/10.3389/fgene.2017.00084 - Bersanelli M et al. Methods for the integration of multi-omics data: mathematical aspects. BMC Bioinformatics 17(2), S15 (2016).
https://doi.org/10.1186/s12859-015-0857-9 - Fondi M and Liò P. Multi -omics and metabolic modelling pipelines: Challenges and tools for systems microbiology. Microbiol Res 171, 52-64 (2015).
https://doi.org/10.1016/j.micres.2015.01.003 - Meng C et al. A multivariate approach to the integration of multi-omics datasets. BMC Bioinformatics 15(1), 162 (2014).
https://doi.org/10.1186/1471-2105-15-162 - Meng C et al. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief Bioinformatics 17(4), 628-641 (2016).
https://doi.org/10.1093/bib/bbv108 - Berger B, Peng J, Singh M. Computational solutions for omics data. Nat Rev Genet 14(5), 333-346 (2013).
https://doi.org/10.1038/nrg3433 - Castro-Vega LJ et al. Multi-omics analysis defines core genomic alterations in pheochromocytomas and paragangliomas. Nat Commun 6(1), 6044 (2015).
https://doi.org/10.1038/ncomms7044 - Karnovsky A et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 28(3), 373-380 (2012).
https://doi.org/10.1093/bioinformatics/btr661 - Guha N et al. A Multi-omics Approach to Identify Biomarkers of Clinically Advanced Retinoblastoma for Diagnostics and Therapeutic Applications. FASEB J 29, 417.7 (2015).
- Aure MR et al. Identifying In-Trans Process Associated Genes in Breast Cancer by Integrated Analysis of Copy Number and Expression Data. PLOS ONE 8(1), e53014 (2013).
https://doi.org/10.1371/journal.pone.0053014 - Chaudhary K et al. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res 24(6), 1248-1259 (2018).
https://doi.org/10.1158/1078-0432.CCR-17-0853 - Yuan Y, Savage RS, Markowetz F. Patient-Specific Data Fusion Defines Prognostic Cancer Subtypes. PLOS Comput Biol 7(10), e1002227 (2011).
https://doi.org/10.1371/journal.pcbi.1002227 - Liang M et al. Integrative Data Analysis of Multi-Platform Cancer Data with a Multimodal Deep Learning Approach. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 12(4), 928-937 (2015).
https://doi.org/10.1109/TCBB.2014.2377729 - Barrett T et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res 41(D1), D991-D995 (2012).
https://doi.org/10.1093/nar/gks1193 - Uhlen M et al. Towards a knowledge-based human protein atlas. Nat Biotechnol 28(12), 1248-1250 (2010).
https://doi.org/10.1038/nbt1210-1248 - Zhang AD, Dai SX Huang JF. Reconstruction and analysis of human kidney-specific metabolic network based on omics data. BioMed Res Int 2013, 1-11 (2013).
https://doi.org/10.1155/2013/187509 - Klein J et al. The KUPKB: a novel Web application to access multiomics data on kidney disease. FASEB J 26(5), 2145-2153 (2012).
https://doi.org/10.1096/fj.11-194381 - Fernandes M and Husi H. FP222 The Chronic Kidney Disease Database (CKDdb). 30((Suppl 3)), iii141 (2015).
https://doi.org/10.1093/ndt/gfv173.04 - Papadopoulos T et al. Omics databases on kidney disease: where they can be found and how to benefit from them. Clin Kidney J 9(3), 343-352 (2016).
https://doi.org/10.1093/ckj/sfv155 - Sanchez-Nino M et al. Albumin-induced apoptosis of tubular cells is modulated by BASP1. Cell Death Dis 6(2), e1644-e1644 (2015).
https://doi.org/10.1038/cddis.2015.1 - Husi H et al. Proteome-Based Systems Biology Analysis of the Diabetic Mouse Aorta Reveals Major Changes in Fatty Acid Biosynthesis as Potential Hallmark in Diabetes Mellitus-Associated Vascular Disease. Circ Cardiovasc Genet 7(2), 161-170 (2014).
https://doi.org/10.1161/CIRCGENETICS.113.000196 - Khan MM et al. Multi-Omics Strategies Uncover Host-Pathogen Interactions. ACS Infect Dis 5(4), 493-505 (2019).
https://pubs.acs.org/doi/10.1021/acsinfecdis.9b00080 - Lee MN et al. Common Genetic Variants Modulate Pathogen-Sensing Responses in Human Dendritic Cells. Science 343(6175), 1246980 (2014).
https://doi.org/10.1126/science.1246980 - Miller M et al. Mapping of Interactions between Human Macrophages and Staphylococcus aureus Reveals an Involvement of MAP Kinase Signaling in the Host Defense. J Proteome Res 10(9), 4018-4032 (2011).
https://doi.org/10.1021/pr200224x - Rebollar EA et al. Using “Omics” and Integrated Multi-Omics Approaches to Guide Probiotic Selection to Mitigate Chytridiomycosis and Other Emerging Infectious Diseases. Front Microbiol 7(68) (2016).
https://doi.org/10.3389/fmicb.2016.00068 - Zhou W et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 569(7758), 663-671 (2019).
https://doi.org/10.1038/s41586-019-1236-x - Le Clerc S, Limou S Zagury JF. Large-Scale “OMICS” Studies to Explore the Physiopatholgy of HIV-1 Infection. Front Genet 10(799) (2019).
https://doi.org/10.3389/fgene.2019.00799 - Pagidipati NJ and Gaziano TA. Estimating Deaths From Cardiovascular Disease: A Review of Global Methodologies of Mortality Measurement. Circulation 127(6), 749-756 (2013).
https://doi.org/10.1161/CIRCULATIONAHA.112.128413 - Deloukas P et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 45(1), 25-33 (2013).
- Leon-Mimila P, Wang J, Huertas-Vazquez A. Relevance of Multi-Omics Studies in Cardiovascular Diseases. Front Cardiovasc Med 6, 91 (2019).
https://doi.org/10.3389/fcvm.2019.00091 - Lau E et al. Integrated omics dissection of proteome dynamics during cardiac remodeling. Nate Commun 9(1), 120 (2018).
https://doi.org/10.1038/s41467-017-02467-3 - Asl HF et al. Expression Quantitative Trait Loci Acting Across Multiple Tissues Are Enriched in Inherited Risk for Coronary Artery Disease. Circ Cardiovasc Genet 8(2), 305-315 (2015).
https://doi.org/10.1161/CIRCGENETICS.114.000640 - Feng Q et al. Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci Rep 6(1), 22525 (2016).
https://doi.org/10.1038/srep22525 - Meder B et al. Epigenome-Wide Association Study Identifies Cardiac Gene Patterning and a Novel Class of Biomarkers for Heart Failure. Circulation 136(16), 1528-1544 (2017).
- Talukdar HA et al. Cross-Tissue Regulatory Gene Networks in Coronary Artery Disease. Cell Syst 2(3), 196-208 (2016).
https://doi.org/10.1016/j.cels.2016.02.002 - Zhang X et al. Novel omics technologies in nutrition research. Biotechnol Adv 26(2), 169-176 (2008).
https://doi.org/10.1016/j.biotechadv.2007.11.002 - Griffin JL et al. An integrated reverse functional genomic and metabolic approach to understanding orotic acid-induced fatty liver. Physiol Genomics 17(2), 140-149 (2004).
https://doi.org/10.1152/physiolgenomics.00158.2003 - Schnackenberg LK et al. An Integrated Study of Acute Effects of Valproic Acid in the Liver Using Metabonomics, Proteomics, and Transcriptomics Platforms. OMICS: A Int J Integr Biol 10(1), 1-14 (2006).
https://doi.org/10.1089/omi.2006.10.1 - Mutch DM et al. An integrative metabolism approach identifies stearoyl-CoA desaturase as a target for an arachidonate-enriched diet. FASEB J 19(6), 599-601 (2005).
https://doi.org/10.1096/fj.04-2674fje - Valdes AM, Glass D, Spector TD. Omics technologies and the study of human ageing. Nat Rev Genet 14(9), 601-607 (2013).
https://doi.org/10.1038/nrg3553 - Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 11(3), 191-203 (2010).
https://doi.org/10.1038/nrg2732 - Gonzalez-Covarrubias V et al. Lipidomics of familial longevity. Aging Cell 12(3), 426-434 (2013).
https://doi.org/10.1111/acel.12064 - Wood SH et al. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. AGE 35(3), 763-776 (2013).
https://doi.org/10.1007/s11357-012-9410-1 - de Magalhães JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25(7), 875-881 (2009).
https://doi.org/10.1093/bioinformatics/btp073 - Ritchie MD et al. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet 16(2), 85-97 (2015).
https://doi.org/10.1038/nrg3868 - Misra BB et al. Integrated omics: tools, advances and future approaches. J Mol Endocrinol 62(1), R21 (2019).
https://doi.org/10.1530/JME-18-0055 - Hernández-de-Diego R et al. PaintOmics 3: a web resource for the pathway analysis and visualization of multi-omics data. Nucleic Acids Res 46(W1), W503-W509 (2018).
https://doi.org/10.1093/nar/gky466 - Vasaikar SV et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 46(D1), D956-D963 (2017).
https://doi.org/10.1093/nar/gkx1090 - Röder B et al. web-rMKL: a web server for dimensionality reduction and sample clustering of multi-view data based on unsupervised multiple kernel learning. Nucleic Acids Res 47(W1), W605-W609 (2019).
https://doi.org/10.1093/nar/gkz422 - List M et al. KeyPathwayMinerWeb: online multi-omics network enrichment. Nucleic Acids Res 44(W1), W98-W104 (2016).
https://doi.org/10.1093/nar/gkw373 - Darzi Y et al. iPath3.0: interactive pathways explorer v3. Nucleic Acids Res 46(W1), W510-W513 (2018).
https://doi.org/10.1093/nar/gky299 - Ghandikota S, Hershey GK, Mersha TN. GENEASE: real time bioinformatics tool for multi-omics and disease ontology exploration, analysis and visualization. Bioinformatics 34(18), 3160-3168 (2018).
https://doi.org/10.1093/bioinformatics/bty182 - Kuo TC, Tian TF, Tseng YJ. 3Omics: a web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst Biol 7(1), 64 (2013).
https://doi.org/10.1186/1752-0509-7-64 - Rohart F, Gautier B, Singh A, Lê Cao KA. MixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol 13(11), e1005752 (2017).
https://doi.org/10.1371/journal.pcbi.1005752
All site content, except where otherwise noted, is licensed under a Creative Commons Attribution 4.0 License