OPEN-ACCESS PEER-REVIEWED
REVIEW
Xiaoshuang Yan, Chaochi Yeh, Linglong Zou*
Bioanalytical Sciences, Shanghai Henlius Biotech, Shanghai, China.
Journal of Applied Bioanalysis. Vol.6. No.3. pages 107-130 (2020).
Published 15 August 2020. https://doi.org/10.17145/jab.20.013 | (ISSN 2405-710X).
*Correspondence: Zou L Bioanalytical Sciences Shanghai Henlius Biotech, Inc, 9/F, Innov Tower, Zone A, No. 1801 Hongmei Road, Xuhui District, Shanghai, China. Phone: +86 2133 395 775.
Citation:
Yan X, Yeh C, Zou L. Clinical Applications of Circulating Tumor DNA, Circulating Tumor Cells, and Exosomes as Liquid Biopsy-Based Tumor Biomarkers. J Appl Bioanal 6(3), 107-130 (2020).
Open-access and Copyright:
©2020 Yan X et al. This article is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Funding/Manuscript writing assistance:
The authors have no financial support or funding to report and they also declare that no writing assistance was utilized in the production of this article.
Competing interest:
The authors have declared that no competing interest exist.
Article history:
Received: 01 April 2020, Revised 23 June 2020, Accepted 27 June 2020.
Abstract
Along with improved knowledge of cancer biology and biotechnical progress, the diagnostic approaches have evolved from tissue biopsies to liquid biopsies. As they provide a minimally invasive tumor detection, liquid biopsies allow early diagnosis and serial assessments of tumor progression.
Discovery and use of circulating tumor markers circulating tumor DNA (ctDNA), circulating tumor cells (CTC), and exosomes have largely expanded the possibility of early diagnosis of cancer, patient stratification, as well as developing a personalized treatment. Based on these circulomes, liquid biopsies can be developed, but each type of liquid biopsies has its own merits and limitations. While ctDNA-based methods represent the most advanced techniques, sensitivity improvement is expected given the rarity of ctDNA in circulation. As intact cancer cells, CTC provide information on cancer cells. However, current CTC capturing procedures are still lack of efficiency. Exosomes are abundant, but they are highly heterogeneous and there is a lack of specific markers for identification. Future efforts are needed to improve operational parameters and clinical performance of each method. Prior to a broad use in clinical settings, it is crucial to standardize the procedure for the specific liquid biopsy method and validate the test with adequate specificity and sensitivity for clinical applications.
Introduction
Targeted therapies and immunotherapies have achieved a great success in treating cancer patients compared to conventional therapies such as chemotherapies. However, the treatment outcome largely depends on expression level of respective targets and immune checkpoints, which are usually assessed with a tissue biopsy. While tissue biopsies represent the gold standard for cancer diagnosis, frequent tissue biopsies are often impractical to perform due to invasiveness of the procedure and the risk of disease spreading it may incur [1,2]. Moreover, the characteristics of cancer tissue can evolve during disease progression, the evidence acquired from tissue biopsies is, therefore, a limited snapshot of specific lesion at a specific time [3]. This limitation leads to incomplete information [4], possibly resulting in errors for tumor diagnosis and/or treatment decision [5].
In contrast to tissue biopsies, liquid biopsies usually use blood specimens that are much easier to collect and can be used to enrich blood-derived circulomes such as circulating tumor DNA (ctDNA), circulating tumor cells (CTC), and exosomes. These circulomes have been found to be related to tumorgenesis and thereby they could be important tools for tumor identification and treatment follow-up. ctDNA is part of cell free DNA (cfDNA). In 1989, cfDNA carrying neoplastic characteristics mutations was reported in plasma of cancer patients [6]. These circulating DNA are actually ctDNA. Since then, tumor-derived alterations, including specific gene mutations, epigenetic alterations, and copy number variations (CNV) have been broadly observed in ctDNA of cancer patients. Several commercial ctDNA detection kits have been approved for cancer diagnosis, such as cobas EGFR mutation test v2 (Roche) and Epi proColon (Epigenomics), both of which are elaborately discussed in ctDNA section [7-9]. Similarly, CTC are also shed by tumors and have been described in patients of several cancer types, mainly breast, colorectal, and prostate cancers. CTC count of ≥ 5 CTC in 7.5 mL of blood has been demonstrated as a reliable prognostic tool for these cancers. CTC cluster was reported to have higher metastatic potential [10]. Now CTC detection has been incorporated into National Comprehensive Cancer Network (NCCN) clinical practice guideline (2017, v3) and American Joint Committee on Cancer (AJCC) staging manual (2018, 8th edition) for metastatic breast cancer diagnosis [11]. Unlike ctDNA and CTC, exosomes are abundant in biological fluids. They also closely correlate with tumors and are elevated in cancer patients compared to healthy controls. Moreover, exosomal proteins are significantly higher in cancer patients at advanced stages than those at early stages. The correlation between exosomal proteins and clinical outcomes could serve as predictive and prognostic biomarkers as protein levels of exosomes changed during and after chemotherapy [12]. Additionally, tumor derived exosomes (TDE) are able to induce oncogenic transformation of normal cells [13]. Thus, ctDNA, CTC and exosomes could serve as potential biomarkers for various clinical applications. This review focuses on the isolation procedures and the clinical applications of these circulating biomarkers for cancer diagnosis, tumor progression monitoring, patient selection, and evaluation of treatment effectiveness.
Tables
[click to enlarge]
ctDNA
Cancer cells have long been known for harboring mutations of genes essential for cell growth control. For example, it was reported that in breast and colon cancers, almost every cancer cell contains approximately 80 mutated genes on average [14]. The ctDNA are tumor-derived DNA fragments found in blood. The typical length of ctDNA ranges from 80 to 200 bps and peaks at 160-180 bps, which is a bit shorter than majority of cfDNA [15]. While mechanisms of ctDNA formation are not fully understood, apoptosis and necrosis of cancer cells are considered as major sources of ctDNA [16]. Active secretion of ctDNA by the cells in tumor tissue can be another mechanism [17].
The abundance of ctDNA was estimated to be less than 0.01% of total circulating DNA or cfDNA, equivalent to less than 10 ng/mL of plasma [18,19]. Due to the rarity of ctDNA in blood, efficient enrichment and sensitive detection are necessary for many applications of ctDNA. A variety of nucleic acid isolation kits are commercially available for fast and convenient isolation of cfDNA. Two approaches, PCR-based and NGS-based methods, are primarily available for detection of gene mutations in ctDNA. While the former is usually used to detect small number of gene mutations, the latter is employed for detection of gene mutations of a large panel. Nowadays, with advent of sensitive digital polymerase chain reaction (PCR) technology, one can quantify as little as 0.001% ~ 0.01% mutated alleles in ctDNA samples [20]. In addition to these PCR-based approaches, a recently available technology named single molecular array (SIMOA) may provide another tool for ctDNA detection. It is based on direct detection of single molecules of DNA on magnetic beads. Briefly, ctDNA samples are subjected to denature to become single-strand DNAs, which are subsequently captured by magnetic beads through hybridization with specific probes (complementary sequences) attached to the beads. After hybridization with biotin-labeled probes, the complexes are incubated with streptavidin-ß-galactosidase for detection. Although SIMOA technology is usually used for protein analysis, it is supersensitive in DNA detection with a reported limit of detection of 0.07 fM [21], providing an attractive alternative to the methods that rely on DNA amplification.
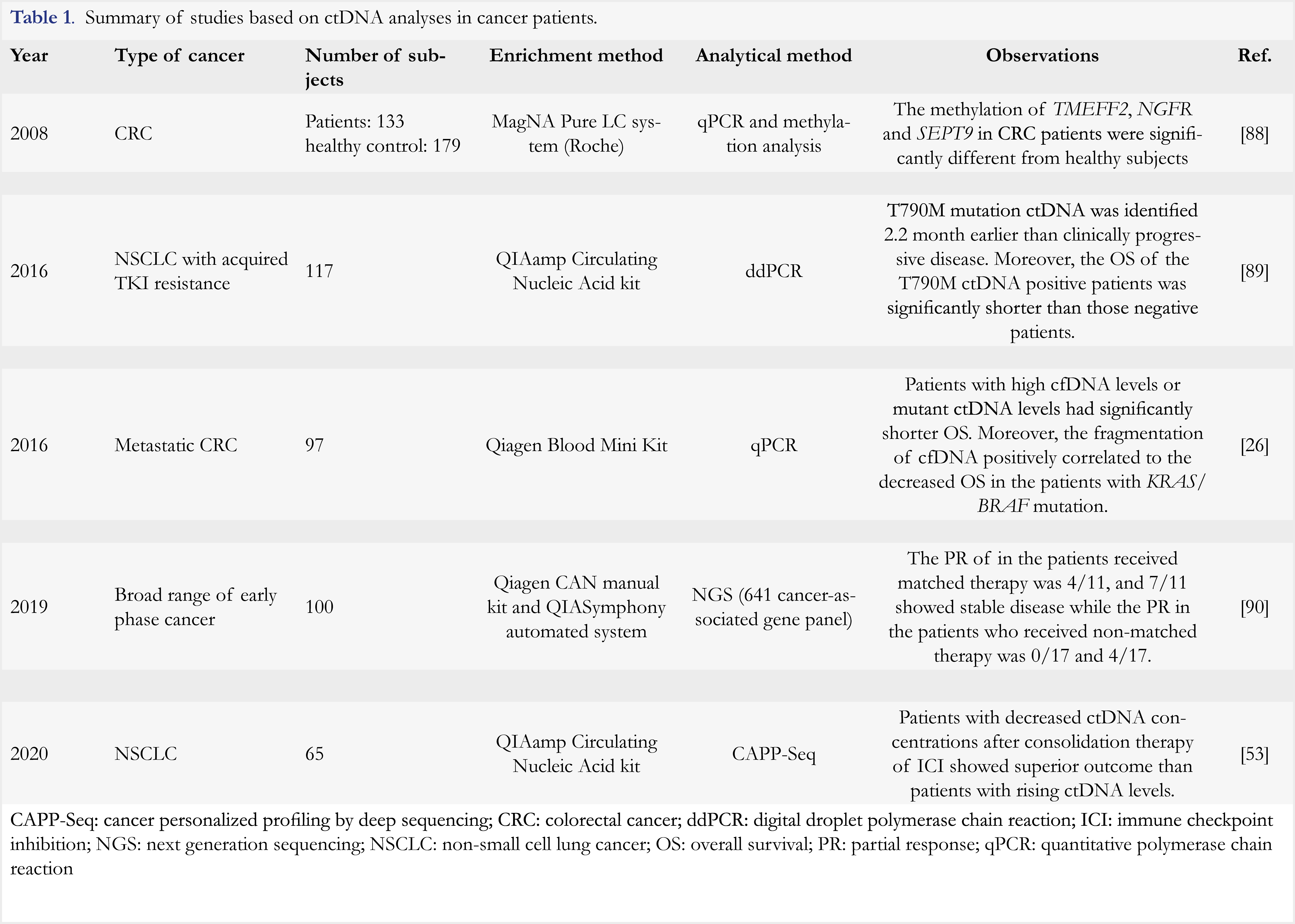
Along with discovery of tumor-driven gene mutations such as EGFR, BRAF, KRAS, and P53, ctDNA analysis has been extensively explored for cancer diagnosis and clinical response monitoring [22, 23]. It was reported that a panel of 16 genes, including but not limited to EGFR, BRAF, KRAS, and NRAS, can be collectively used to detect eight common cancer types [24]. In a study of metastatic melanoma patients treated with immunotherapy, mutations of genes BRAF, NRAS, TERT and ALK were found in ctDNA samples from five of ten (50%) patients. Moreover, three patients with disease progression exhibited an increase in ctDNA levels [25]. In another study of breast cancer [26], ctDNA levels were reported to correlate with degree of tumor burden better than CA15-3, a typical protein biomarker used for monitoring breast cancer. ctDNA also displayed a better performance in monitoring the treatment response in this study. In addition to its potential applications in cancer diagnosis and treatment monitoring, ctDNA analysis of tumor-driven genes mutations can be used to predict drug resistance and cancer recurrence. For instance, dynamic ctDNA profiling of genes TP53, PIK3CA, mTOR, and Pten in HER2-positive breast cancer patients could be used to identify those resistant to anti-HER1/2 tyrosine kinase inhibitor therapy with sensitivity of 85.7% [27]. Additionally, simultaneous analysis of KRAS, TP53, PIK3CA and APC gene mutations in ctDNA samples from colorectal cancer patients could highly predict the recurrence of cancer within one year [28]. In colorectal cancer patients after surgical resection, postoperative gene mutations of KRAS, APC and P53 detected in ctDNA samples indicated a high risk of recurrence, especially in patients without the adjuvant chemotherapy treatment (79% versus 9.8% at median follow-up of 27 months) [29]. All of these results indicated that ctDNA could serve as a promising biomarker for patient selection, treatment evaluation and recurrence monitoring, as well as drug resistance prediction. Clinical uses of ctDNA analyses were reported in various studies and some of them are summarized in Table 1.
Currently, there are two ctDNA-based tests approved by Food and Drug Administration (FDA). The first one was approved in 2016 as a companion diagnostic for detection of EGFR mutation in patients with non-small cell lung cancer (NSCLC) (Cobas® EGFR Mutation Test v2, Roche). This test is to select the patients who would benefit from erlotinib and osimertinib treatment [7,8]. It is based on an optimized quantitative PCR technology using pre-amplification to improve the detection efficiency of mutated alleles. The clinical specificity and sensitivity of this test in NSCLC were 97.9% and 72.1%, respectively. Compared with tissue biopsy-based assay, the cobas® EGFR Mutation Test v1, the concordance between ctDNA and tissue biopsy analyses was 91% [30]. Another approved diagnostic test is Epi proColon (Epigenomics). This test is used for colorectal cancer diagnosis and it is based on the detection of ctDNA methylation [31]. Investigation of tumor epigenetics, including methylation and histone post-transcriptional modifications, represent important applications of ctDNA analysis [32]. In recent years, several studies have been conducted to analyze ctDNA methylation patterns for cancer screening, early diagnosis, and follow-up of the disease progression [33, 34]. Some examples in this application are shown in Table 1. Several NGS-based ctDNA tests have been marketed as laboratory-developed tests (LDT) since 2014, such as Guardant360® (Guardant Health, 2014), FoundationAct (Foundation Medicine, 2016), Oncotype SEQ (Genomic Health, 2016), and PlasmaSELECT (Personal Genome Diagnostics, 2015). They are used to detect four major types of gene alterations, including point mutation, insertions/deletions, copy number changes, and gene fusions. Normally offered in the Clinical Laboratory Improvement Amendments (CLIA)-regulated and College of American Pathology (CAP)-accredited laboratories, these tests have shown good performance. As an example, Guardant360® exhibits clinical sensitivity of 85% and specificity of 99.9% in advanced stage solid tumors, and the analytical sensitivity and specificity of this test were 100% and 99.99%, respectively [9]. For further information about these LDT, readers can refer to review article [35].
While ctDNA analysis has shown a great potential in various clinical applications, it exhibits several challenges. First, it is difficult to obtain good quantity of ctDNA as its content in peripheral blood is extremely low (< 0.01% of 10 ng/mL plasma). Moreover, half-lives of ctDNA are reported to be 16 minutes to several hours [28], which are short and may partially explain low abundance of ctDNA. Second, ctDNA detection platforms can vary from genome-wide analysis to a single gene interrogation. These analyses exhibit different detection limits (0.1-1.0% for allele-specific PCR, 0.01-2.0% for NGS, and 0.01% for digital PCR and cancer personalized profiling by deep sequencing (CAPP-Seq)) [36]. Therefore, analysis results can vary with different platform technologies, rendering evaluation of results difficult among different laboratories that employ different platforms. Another challenge lies in poor concordance between liquid biopsies and tissue biopsies in certain cases. As an example, the concordance of EGFR L858R and 19del mutation between ctDNA- and tissue biopsy-based analyses in NSCLC patients was 91%, while it was only 61% for T790M mutation [37]. This might be attributable to different disease stages, as high concordance between liquid and tissue biopsies of PIK3CA mutation status was observed in advanced breast cancer patients, but a poor concordance was reported in patients at early-stage [38]. Combined analyses of other biomarkers with ctDNA samples could be used to improve the clinical value of ctDNA-based liquid biopsies. An example was provided by Cohen et al, who reported that through a combined analysis of eight protein biomarkers and ctDNA markers, 8 different cancer types could be identified with specificity of >99%, and sensitivity of 69% to 98%, depending on cancer type [24]. Additionally, a combined analysis of DNA from both exosomes and CTC might be an alternative to obtain more reliable results in clinical applications [39].
CTC
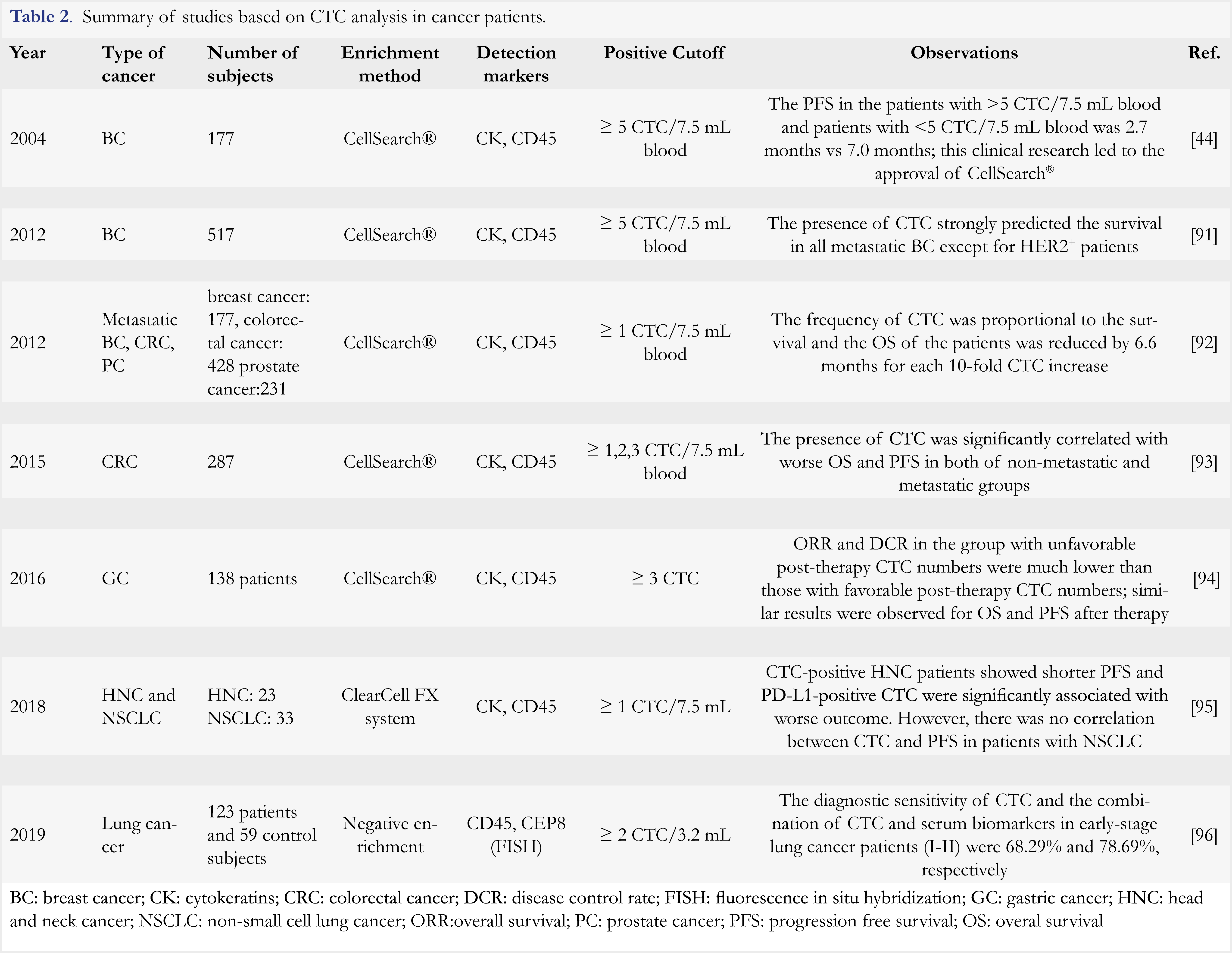
CTC are tumor cells that detached from primary and metastatic tumor lesions and enter into blood circulation. Since their discovery in 1869, CTC have been widely identified in patients with breast cancer [40], non-small cell lung cancer [41], prostate cancer [42], colon cancer [43], and pancreatic cancer [44]. As intact cells derived from tumor, CTC can provide valuable information on tumor composition, heterogeneity, invasiveness, and drug resistance. As early as in 2004, it was found in a clinical study conducted in breast cancer patients that the number of CTC was inversely correlated to progression-free survival (PFS) and overall survival (OS) of the patients receiving an anti-cancer treatment [45]. Similar findings were also reported in patients with prostate cancer and patients with colorectal cancer (CRC) [46,47]. In patients with castration-resistant prostate cancer, Danila et al reported that the 2-year OS was 46% in the patients with low level of CTC and 2% in the patients with high level of CTC, indicating a prognostic value of CTC [48]. More studies on the clinical applications of CTC enumeration are listed in Table 2.
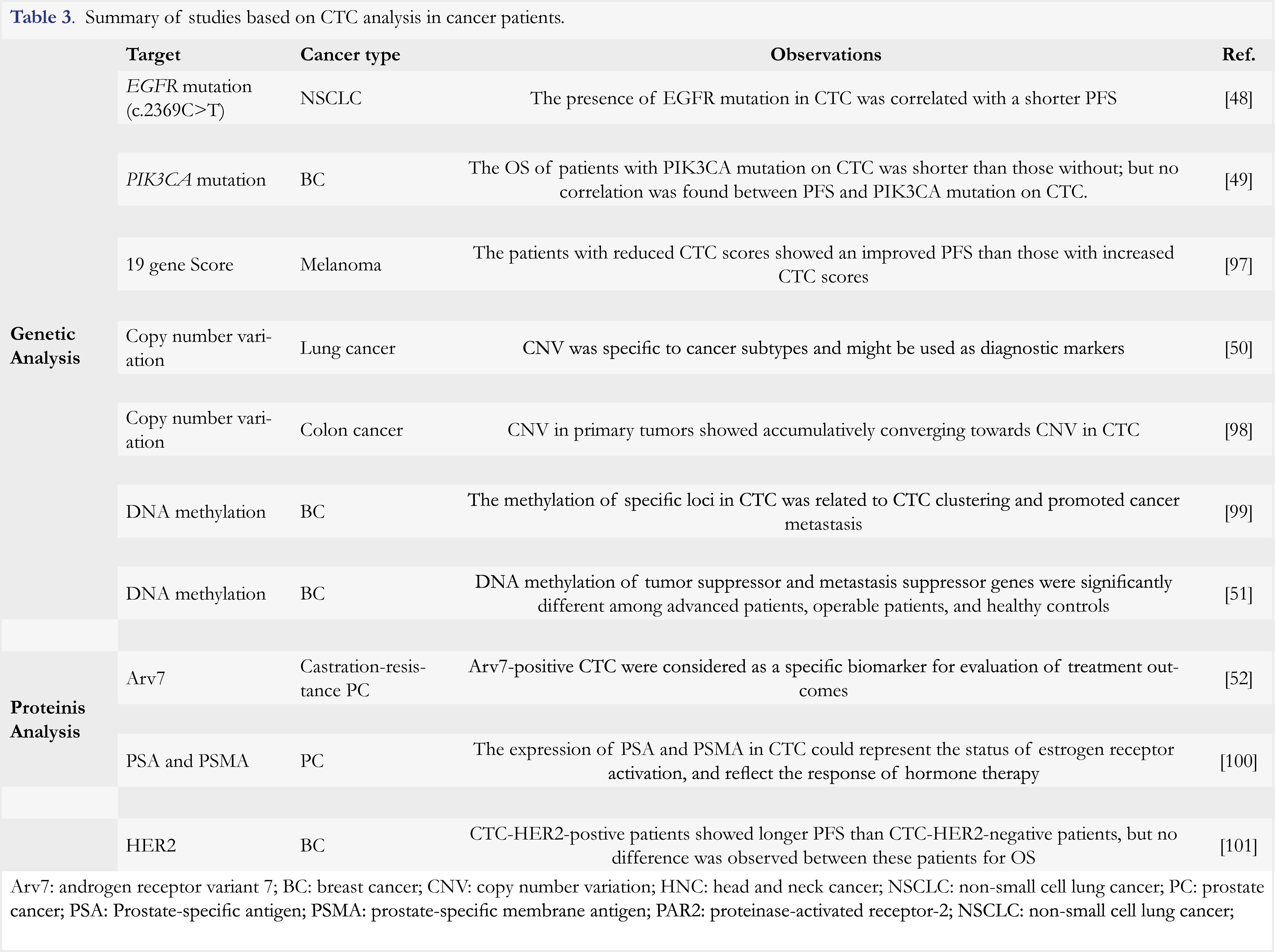
In addition to CTC enumeration, genetic and protein analyses of CTC also offer a potential value for cancer diagnosis and prognosis. The commonly studied tumor-driven gene mutations were reported in CTC. For example, EGFR and PIK3CA mutations were observed in CTC of NSCLC and breast cancer patients, respectively [49, 50]. Moreover, the presence of EGFR mutation was found to be associated with shorter PFS in NSCLC patients and PIK3CA mutation were related to shorter OS in breast cancer patients. Further molecular analyses are summarized in Table 3. The studies demonstrated a significant value of CTC in cancer diagnosis, prognosis and treatment response evaluation. Indeed, in patients with different cancers, CTC showed distinct pattern of CNV, which was consistent with results of tissue biopsy analysis from metastatic lesions in the same patients [51]. Similar results were also observed for DNA methylation in CTC. As reported by Chimonidou et al [52], the methylation patterns of tumor suppressor and metastasis suppressor genes in CTC were found remarkably distinct in patients versus healthy individuals, and in metastatic patients versus operable patients. All of these findings indicated that CTC could serve not only as a diagnostic biomarker, but also as a prognostic biomarker as they provided valuable information for tracking cancer metastasis. Protein analysis can be performed on CTC in clinical studies as well. A study conducted in patients with castration-resistance prostate cancer revealed androgen receptor variant 7 (Arv7)-positive CTC as specific predictive biomarker for prostate cancer [53]. In another study, expression of prostate-specific antigen (PSA) and prostate-specific membrane antigen (PSMA) was analyzed in CTC [54]. It was found that these two proteins could represent different status of androgen receptor (AR) activation. Moreover, the elevated number of “AR-on” CTC (CTC with PSA+/PSMA– phenotype) was associated with reduction in OS in patients. Recently, the expression of immune checkpoint inhibitors (e.g. PD-L1) and other therapeutic targets such as HER2 was also found in CTC [55]. The expression analysis of these therapeutic targets in CTC showed important clinical value in responsiveness and drug resistance evaluation. As an example, a study in NSCLC patients reported that the number of CTC with higher PD-L1 inversely correlated to clinical response. In other words, the more CTC expressing high level of PD-L1 in the patients, the stronger resistance to anti-PD1 antibody treatment [55].
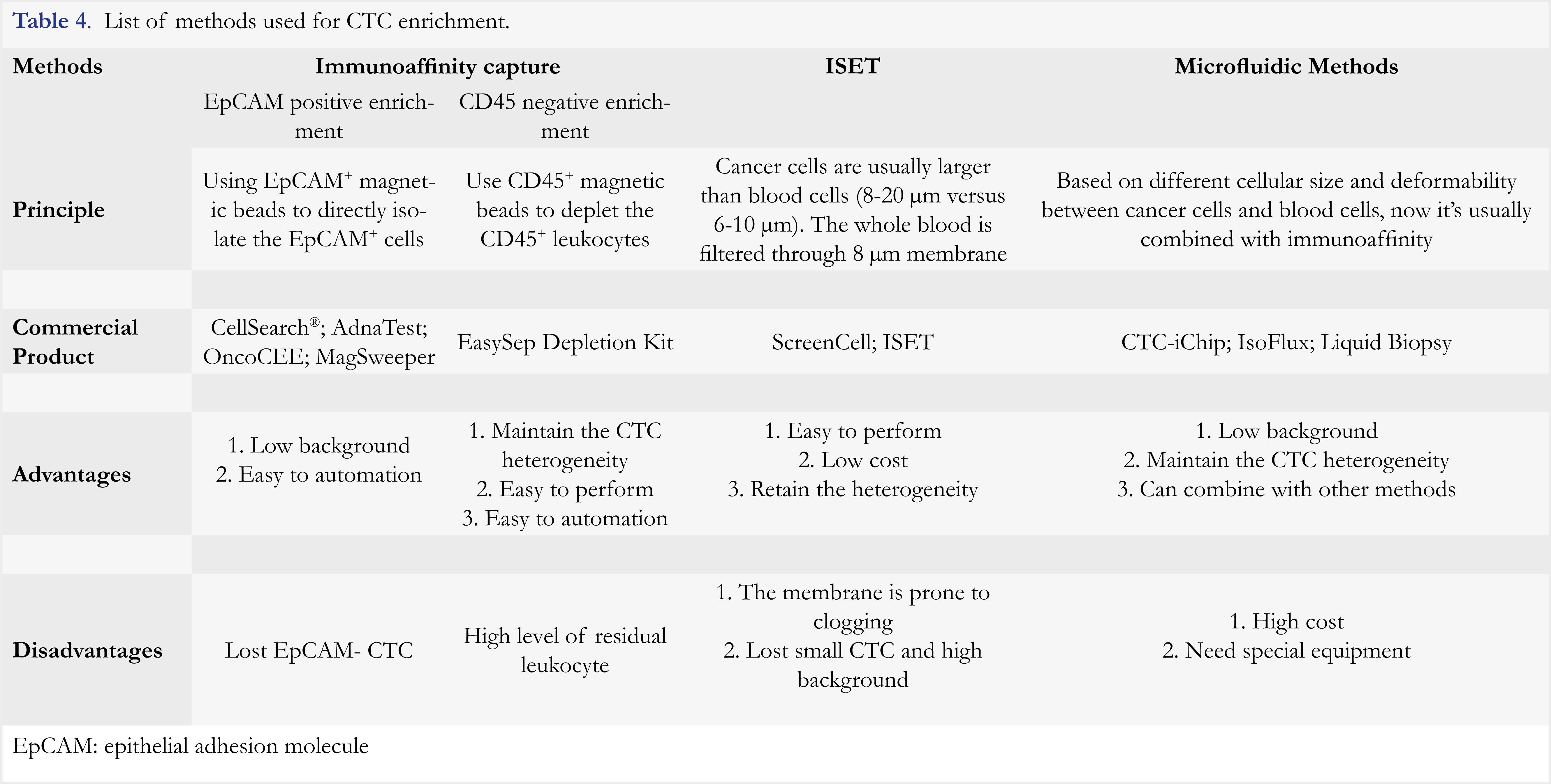
The prevalence of CTC is approximately 5 to 200 CTC in 7.5 mL of blood. Given this rarity, CTC enrichment is critical for its clinical application. There are three strategies employed for CTC enrichment, including immuno-affinity-based capture, isolation by size of epithelial tumor cells (ISET) [56], and microfluidics-based methods [57]. Immunoaffinity-based capture approaches are most widely used, and microfluidics-based methods are usually used in combination with immunoaffinity-based capture [58]. The comparison of these enrichment methods is summarized in Table 4. The interested readers could also refer to several reviews for more information [59, 60]. The most widely used platform for CTC enrichment is an epithelial adhesion molecule (EpCAM)-based immunoaffinity capture assay such as CellSearch® (Veridex). EpCAM is an epithelial cell-specific marker ubiquitously expressed on normal epithelial cells and cancer cells, but not on blood cells, whereas CD45 is a leukocyte-specific marker. Cytokeratins (CK) are also epithelial cell-specific markers and highly expressed on cancer cells. Thus, CTC are defined as CD45– CK+EpCAM+ cells by immunofluorence staining. The cut-off threshold in this method for distinguishing positive from negative results is 5 CTC in 7.5 mL of blood. This method was approved by FDA in 2004 for prognosis of metastatic breast cancer [45]. Until now, it is the only FDA-cleared method for CTC enrichment.
Although the use of CellSearch® has resulted in remarkable progress in CTC detection, this method has inherent drawbacks as it only relies on the expression of EpCAM for CTC enrichment. EpCAM is not a unique biomarker for CTC, and it could be present on normal epithelia cells. On the other hand, a significant number of CTC lacks EpCAM expression in patients with a non-epithelial cancer, such as melanoma [61]. With cumulative knowledge on epithelial-mesenchymal transition (EMT), it becomes clear that mesenchymal CTC would express mesenchymal markers such as Vimentin and Twist [62-64], but not epithelial marker EpCAM. In addition, EpCAM on epithelial CTC could be down-regulated or even lost during CTC circulation [65]. The use of EpCAM for CTC isolation would thereby miss out on these EpCAM-negative CTC. Recently, several subpopulations of CTC have been identified, which include epithelial CTC (E-CTC) that express EpCAM, mesenchymal CTC (M-CTC) that express vimentin, and biphenotypic E/M-CTC (expressing both EpCAM and vimentin), as well as circulating tumor microemboli (CTMs) [66]. Thus, the use of CellSearch® has resulted in low sensitivity (5-30%) of CTC detection [67], as it detects epithelial CTC only. These findings indicate that CTC in peripheral blood are highly heterogeneous, and thereby it is inadequate to identify and characterize CTC based on EpCAM marker only.
Despite encouraging advancements have been achieved in the last decades, several barriers still exist, which prevent wide-spread clinical applications of CTC. First, the rarity of CTC poses a significant challenge to CTC utilization, warranting enrichment of CTC with high purity and quality. Second, CTC are highly heterogeneous and have no universal markers. The identification of CK+EpCAM+CD45– cells as CTC is imprecise as quite a few CTC neither express CK nor EpCAM. Finally, CTC numeration is strongly dependent on the blood volume, enrichment method, and photographic or image processing system [68]. Distinct cutoffs were reported among different methods for distinguishing positive from negative samples (5 CTC/7.5 mL for CellSearch®, 50 CTC/mL for ISET, versus 14 CTC/mL for CTC-chip) [69,70], rendering analytical results from different methods difficult to compare. Thus, the future efforts will be needed to improve current methodologies for expanded use of CTC in the following aspects: 1) Development of more useful isolation method that may combine different isolation mechanisms (e.g. Immunoaffinity-based capture and microfluidics); 2) Establishment of standard isolation method that yields as many CTC as possible without losing their heterogeneity; 3) Identification of more specific markers to CTC, and with more defined criteria to objectively identify CTC; 4) Confirmation of clinical value of CTC as a diagnostic, prognostic or predictive biomarker through large-scale clinical trials. Advances in these aspects would enable the better value of CTC as biomarkers for tumor management.
Exosomes
Exosomes are small lipid bilayer vesicles of 30 to 200 nanometers in diameter [71]. Released from almost every type of eukaryocytes through a sequential process, they are enriched with multiple cargoes of cellular origin, including lipids, proteins, and nucleic acids. Since the initial discovery in sheep reticulocytes in 1983 [72], exosomes have been considered as powerful shuttles for transporting biofunctional cargoes among cells, and have been implicated in both physiological and pathological processes. Studies have indicated that tumor-derived exosomes (TDE) participate in cancer development. As reported by Roccaro et al, the exosomes released from bone marrow-derived mesenchymal cells promoted the multiple myeloma (MM) development in animal models [73]. Similar result was observed in breast cancer cells that TDE could induce oncogenic transformation of normal cells [74]. This could be attributable to the involvement of exosomes in various processes that facilitated tumor progression, for example, angiogenesis, EMT, and drug resistance. Angiogenesis is possibly mediated by activation of the PAR2 signaling, an established angiogenic pathway, by exosomes [75]. The EMT regulators such as TGF-ß, ß-catenin, and tumor necrosis factor alpha (TNFα) are widely found in exosomes and they enhance migratory and invasive capacity of cancer cells [76]. Drug resistance is mediated by TDE possibly via several mechanisms, such as encapsulating and exporting the drugs from cancer cells [77], transferring multi-drug resistance (MDR)-associated proteins into cancer cells, and binding to the drugs and thereby blocking them [78].
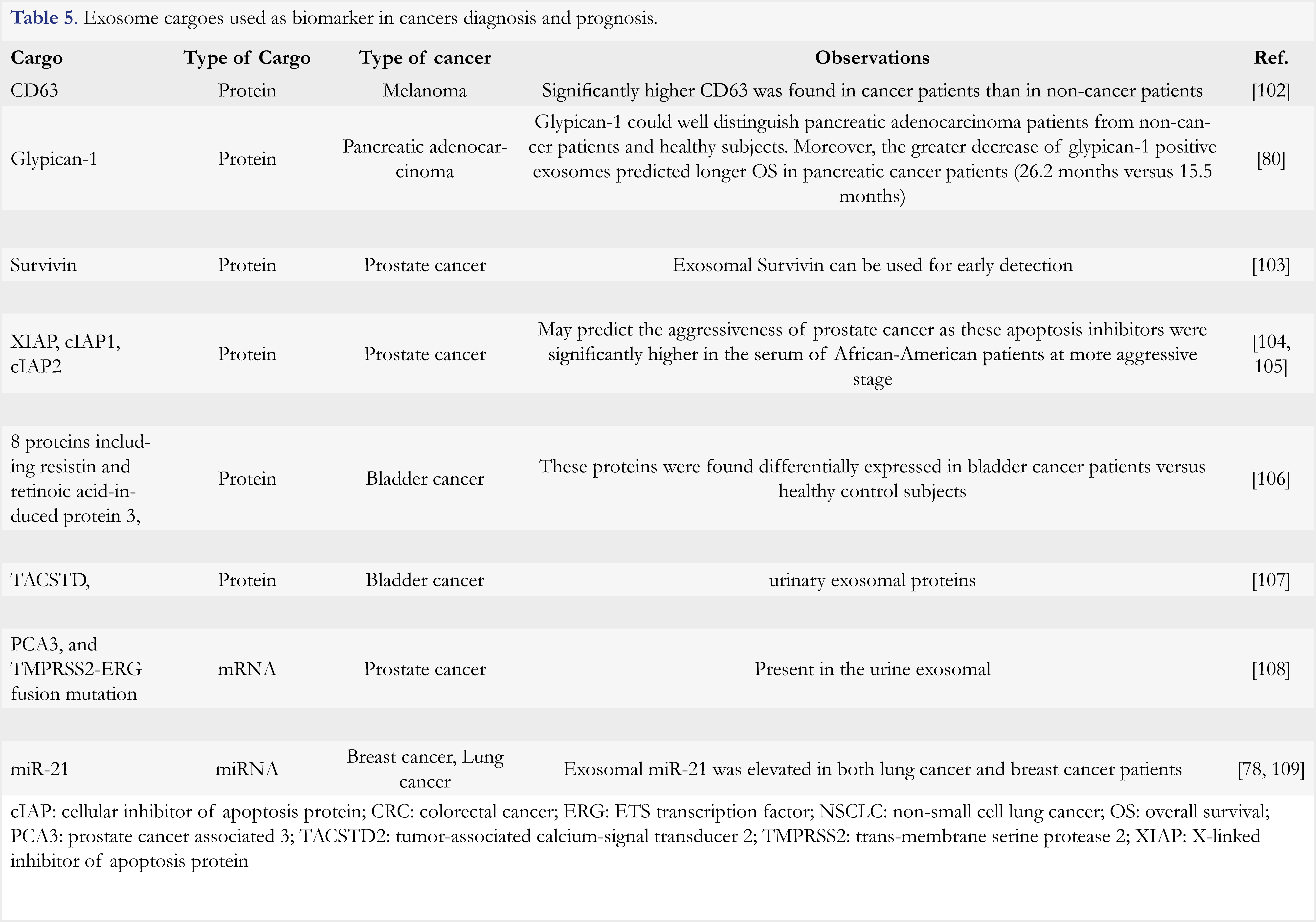
Due to easy access from biological fluids and given their roles in tumor progression and drug resistance, exosomes have been considered as attractive tools for cancer diagnosis and treatment evaluation. A markedly increased release of exosomes was observed in the serum of lung adenocarcinoma patients compared to non-cancer subjects (2.85 mg/mL versus 0.77 mg/mL) [79,80]. Both exosomal proteins and nucleic acids have been explored as biomarkers for clinical use. In a study of pancreatic adenocarcinoma, author revealed that the quantities of glypican-1-positive exosomes could be used to reliably distinguish the adenocarcinoma patients from non-cancer patients and healthy subjects [81]. The messenger RNA of EGFRv III, a common tumorigenic mutation known as an in-frame deletion of exons 2–7 in the coding sequence of EGFR, was found in the exosomes of glioblastoma patients [82].
There are many other cargoes reported in exosomes, which are listed in Table 5. One type of cargoes of exosomes is microRNA (miRNA). Contents of miRNA can be different in exosomes from cancer patients versus healthy subjects. In the aforementioned study of lung adenocarcinoma, the mean miRNA concentration was significantly higher in patients than in control subjects (158.6 ng/mL versus 68.1 ng/mL) [79]. Besides, it was found that various specific miRNA could discriminate cancer patients from healthy subjects. For example, 8 species of miRNA were uniformly elevated in the patients with advanced-stage ovarian tumor, while they remained at much lower levels in benign patients and even absent in healthy controls [83]. Recently, PD-L1 was found to be specifically packaged into exosomes of prostate cancer cells and to be involved in inhibiting T cell activation and promoting cancer cell growth [84]. Moreover, another clinical study of melanoma showed that exosomal PD-L1 was exclusively related to pembrozumab resistance, and the level of exosomal PD-L1 is much higher in non-responders than that in responders [85]. This finding suggested that exosomal checkpoint inhibitor levels could serve as a potentially useful surrogate for predicting the clinical response in immunotherapy.
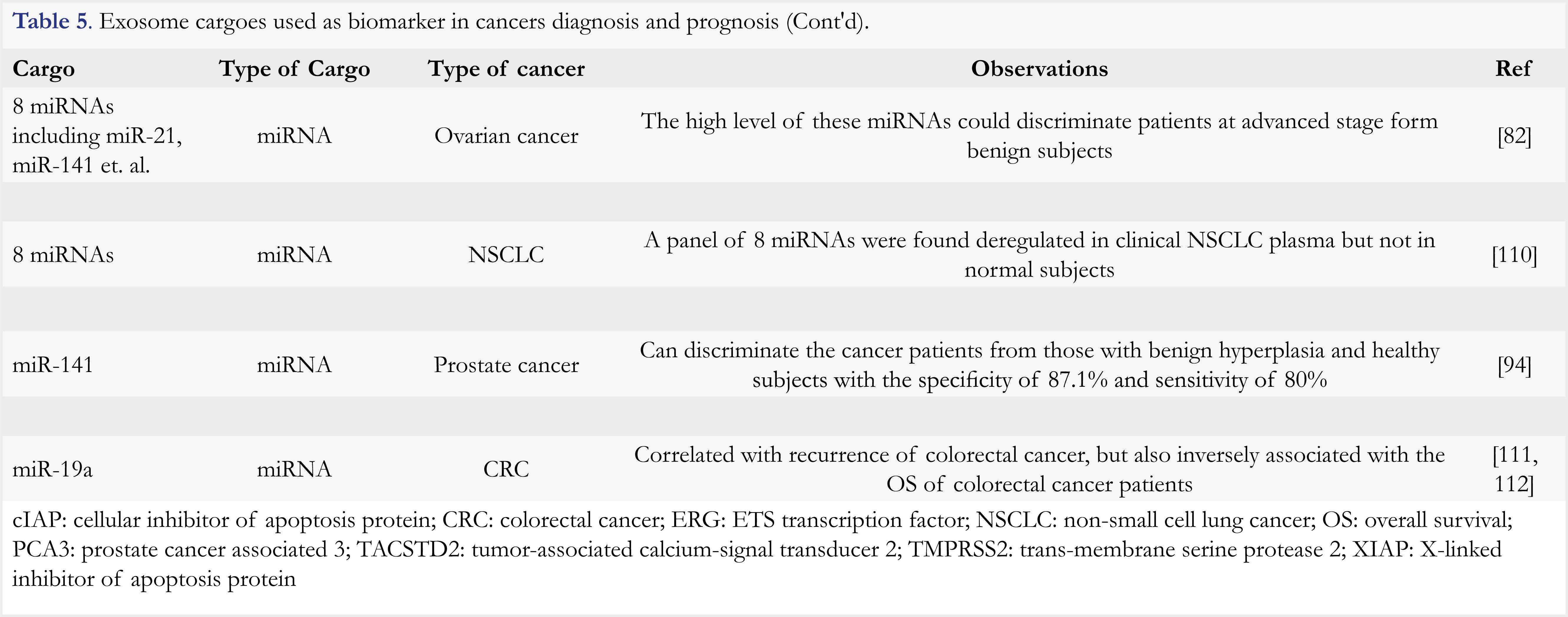
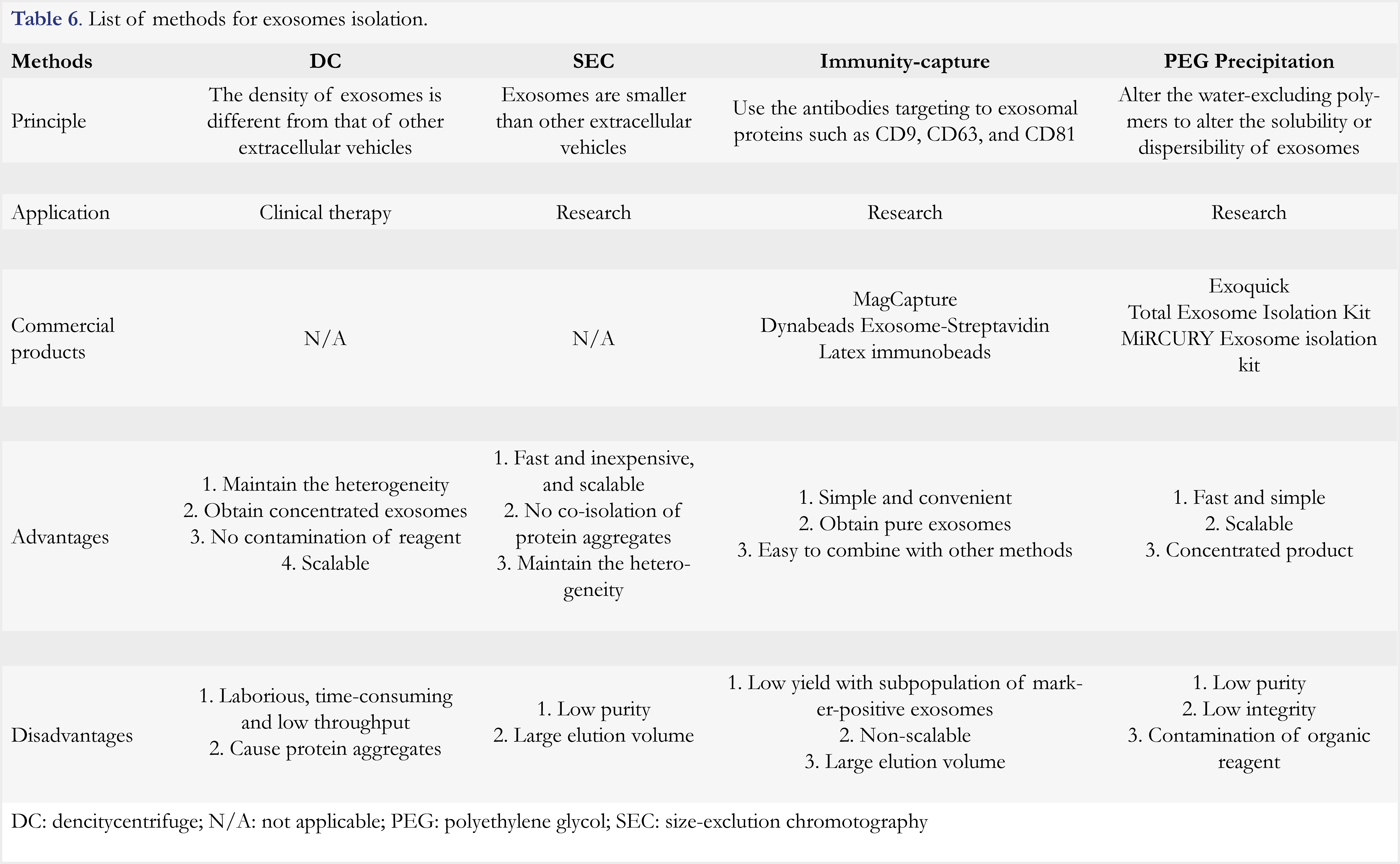
In summary, exosomes are multifunctional entities that play important roles in tumor progression and drug resistance. However, high quality exosomes are difficult to prepare as they are highly heterogeneous in terms of size, cargo content, and cellular origin. Five types of methods have been developed, which include differential centrifugation, size exclusion chromatography, immune-capture, polyethylene glycol precipitation, and microfluidic-based methods. These methods are based on the distinct features of density, size, surface proteins, and hydrophobicity of exosomes, respectively. The principles, applications and commercial products, as well as advantages and disadvantages of each method are provided in Table 6, and in a review article [86]. Despite availability of various methods for exosomes isolation, there is no single perfect method, and therefore methods are usually used in combination. As different laboratories use different methods and/or different markers for cancer identification, the results are hardly comparable among laboratories. Thus, further efforts are needed to standardize the exosomes-isolating method and to identify common markers for use.
Comparison of three circulating tumor markers
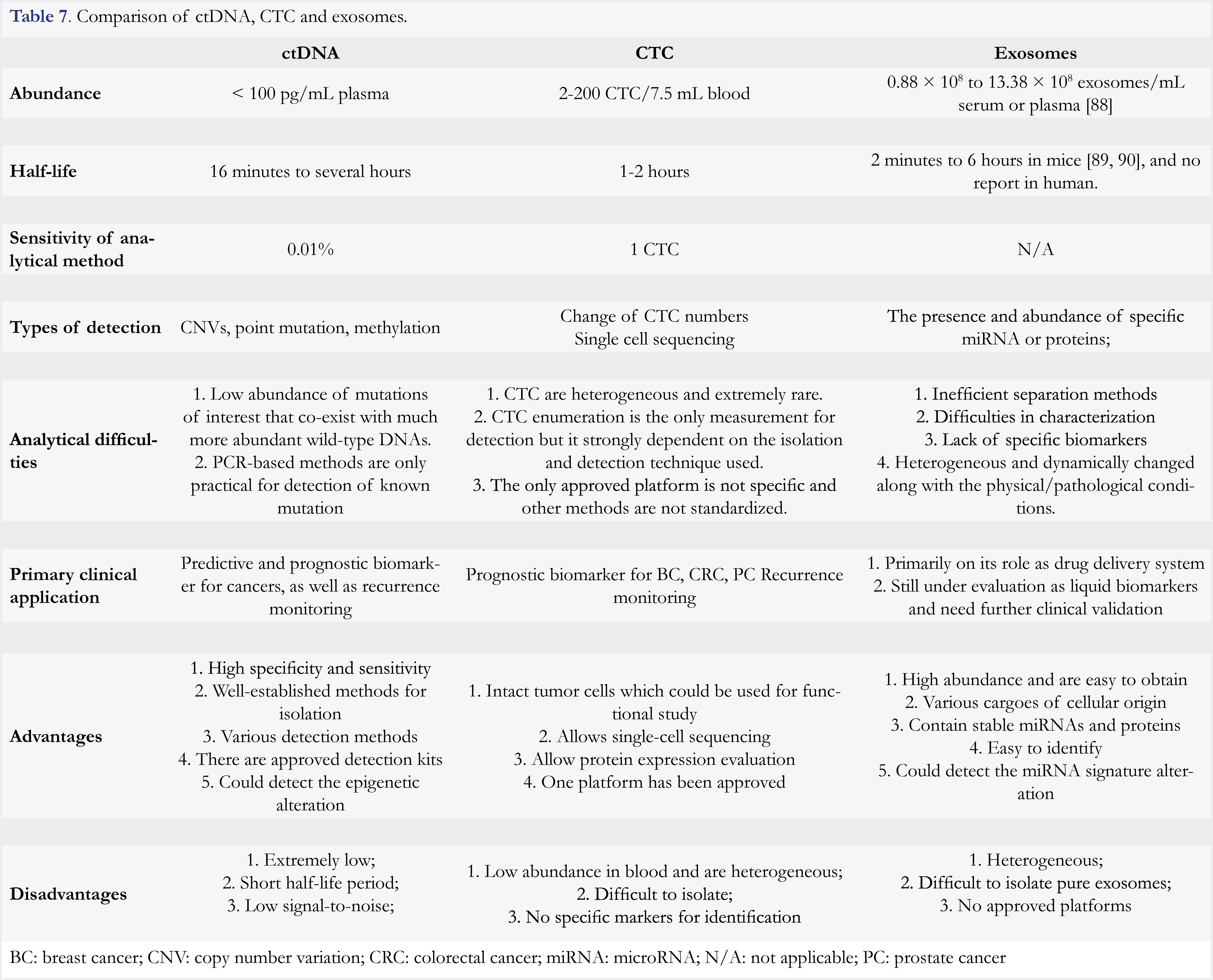
Circulating tumor markers ctDNA, CTC, and exosomes have largely expanded the possibility of early diagnosis of cancer, patient stratification, as well as developing a personalized treatment. Based on these circulomes liquid biopsies can be developed, but each type of liquid biopsies has its own merits and limitations. Among the three circulomes, ctDNA-based methods represent the most advanced techniques. Several commercial products or LDT platforms have been launched. As it detects gene mutations with the sensitivity of 0.01%, a ctDNA test is broadly used for patient selection, medication guidance, and recurrence monitoring. However, it is difficult to distinguish real CNV from an operational error or aging-associated clonal hematopoietic mutations of indeterminate potential [87]. Moreover, PCR-based ctDNA detection is of low throughput and a sequencing-based method is of high cost. CTC is also an approved biomarker for prognosis of breast cancer. By enumeration, CTC is approved as adjunctive diagnosis for breast cancer staging. As intact cancer cells, they can be subjected to single cell analysis at both nucleic acid and protein levels. Furthermore, CTC can be cultivated in vitro for other analyses. However, CTC capture and analysis are costly. In addition, ambiguous correlation between CTC and tumor progression and treatment outcome might complicate cancer diagnosis and prognosis [88]. Exosomes are abundant, which makes them easier to obtain. Moreover, extensive studies have revealed that the miRNA enclosed in exosomes are differentially present in cancer patients versus healthy controls, which holds a great potential for cancer diagnosis. However, exosomes are highly heterogeneous and there is a lack of specific markers for identification. Thus, technologies remain to be further developed before one can effectively apply exosomes in cancer diagnosis. For further comparison among these three types of circulomes, readers can refer to Table 7.
Summary and future perspectives
These circulomes and the associated liquid biopsy methods elaborated above provide promising supplemental tools, and in some cases, the tool alternative to tissue biopsies. Although remarkable advances have been made in last few decades, clinical applications of the liquid biopsies is still challenging due to the rarity and difficulty in enrichment of these circulomes. Future efforts are needed to improve clinical performance of each method. For ctDNA analysis, efforts should be directed to develop high throughput digital PCR instrument with a further improved signal-noise ratio. For CTC analysis, an urgent need would be to develop an immune-affinity enrichment protocol based on multiple antibodies against both epithelial and mesenchymal markers. For exosomes, the focus of efforts should be to identify cancer tissue origin of exosomes and to validate the correlation between exosomes and cancers in large clinical studies. It should be noted that except approved methods, the isolation procedures of circulomes and design of clinical trials vary significantly in different laboratories and clinical centers, resulting in poor comparability. Thus, prior to a broad use in clinical settings, it is crucial to standardize the procedure for the specific liquid biopsy method and validate the test with adequate specificity and sensitivity for clinical applications. Progresses in liquid biopsy technology would yield significant benefit for cancer diagnosis, patient selection, and treatment effect monitoring.
References
- Shyamala K, Girish HC, Murgod S. Risk of tumor cell seeding through biopsy and aspiration cytology. J Int Soc Prev Community Dent 4(1), 5-11 (2014).
https://doi.org/10.4103/2231-0762.129446 - Robertson EG, Baxter G. Tumour seeding following percutaneous needle biopsy: the real story! Clin Radiol 66(11), 1007-1014 (2011).
https://doi.org/10.1016/j.crad.2011.05.012 - Marrugo-Ramirez J, Mir M, Samitier J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int J Mol Sci 19(10), (2018).
https://doi.org/10.3390/ijms19102877 - Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, Mcdonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366(10), 883-892 (2012).
https://doi.org/10.1056/NEJMoa1113205 - Tzeng YT, Chang SE, Mei R, Javey M. Liquid Biopsy Prevents Inaccurate Her2 Status Determination by in situ Hybridization in a Patient with Invasive Ductal Adenocarcinoma of the Breast: Case Report. Case Rep Oncol 10(3), 857-862 (2017).
https://doi.org/10.1159/000480698 - Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 46(5), 318-322 (1989).
https://doi.org/10.1159/000226740 - Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med 5(3), 46 (2017).
https://doi.org/10.21037/atm.2017.01.32 - FDA Grants First Liquid Biopsy Approval to the Roche cobas EGFR Mutation Test v2. (2016). Avaible at https://diagnostics.roche.com/global/en/news-listing/2016/fda-grants-first-liquid-biopsy-approval-to-the-roche-cobas-egfr-mutation-test-v21.html
- Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, Collisson EA, Divers SG, Hoon DS, Kopetz ES, Lee J, Nikolinakos PG, Baca AM, Kermani BG, Eltoukhy H, Talasaz A. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One 10(10), e0140712 (2015).
https://doi.org/10.1371/journal.pone.0140712 - Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, Brannigan BW, Kapur R, Stott SL, Shioda T, Ramaswamy S, Ting DT, Lin CP, Toner M, Haber DA, Maheswaran S. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158(5), 1110-1122 (2014).
https://doi.org/10.1016/j.cell.2014.07.013 - Jessica K, Dohler BS. AJCC cancer staging manual 8th edition Chapters 1&2 summary review (2017). Available at
- Szajnik M, Derbis M, Lach M, Patalas P, Michalak M, Drzewiecka H, Szpurek D, Nowakowski A, Spaczynski M, Baranowski W, Whiteside TL. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet (Sunnyvale) Suppl 4(1), 003-013 (2013).
- Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18(6), 883-891 (2012).
https://doi.org/10.1038/nm.2753 - Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science 318(5853), 1108-1113 (2007).
https://doi.org/10.1126/science.1145720 - Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, Del Rio M, Molina F. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 38(18), 6159-6175 (2010).
https://doi.org/10.1093/nar/gkq421 - Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 61(4), 1659-1665 (2001).
- Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 10(8), 472-484 (2013).
https://doi.org/10.1038/nrclinonc.2013.110 - Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr., Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 102(45), 16368-16373 (2005).
https://doi.org/10.1073/pnas.0507904102 - El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 424(1), 222-230 (2013).
https://doi.org/10.1016/j.cca.2013.05.022 - Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA 96(16), 9236-9241 (1999).
https://pnas.org/doi/full/10.1073/pnas.96.16.9236 - Song L, Shan D, Zhao M, Pink BA, Minnehan KA, York L, Gardel M, Sullivan S, Phillips AF, Hayman RB, Walt DR, Duffy DC. Direct detection of bacterial genomic DNA at sub-femtomolar concentrations using single molecule arrays. Anal Chem 85(3), 1932-1939 (2013).
https://doi.org/10.1021/ac303426b - Formisano L, Lu Y, Servetto A, Hanker AB, Jansen VM, Bauer JA, Sudhan DR, Guerrero-Zotano AL, Croessmann S, Guo Y, Ericsson PG, Lee KM, Nixon MJ, Schwarz LJ, Sanders ME, Dugger TC, Cruz MR, Behdad A, Cristofanilli M, Bardia A, O’shaughnessy J, Nagy RJ, Lanman RB, Solovieff N, He W, Miller M, Su F, Shyr Y, Mayer IA, Balko JM, Arteaga CL. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun 10(1), 1373 (2019).
https://doi.org/10.1038/s41467-019-09068-2 - Hu ZY, Xie N, Tian C, Yang X, Liu L, Li J, Xiao H, Wu H, Lu J, Gao J, Hu X, Cao M, Shui Z, Xiao M, Tang Y, He Q, Chang L, Xia X, Yi X, Liao Q, Ouyang Q. Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients with Multiline Resistance. EBioMedicine 32(1), 111-118 (2018).
https://doi.org/10.1016/j.ebiom.2018.05.015 - Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA, Jr., Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359(6378), 926-930 (2018).
https://doi.org/10.1126/science.aar3247 - Lipson EJ, Velculescu VE, Pritchard TS, Sausen M, Pardoll DM, Topalian SL, Diaz LA, Jr. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer 2(1), 42 (2014).
https://doi.org/10.1186/s40425-014-0042-0 - Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368(13), 1199-1209 (2013).
https://doi.org/10.1056/NEJMoa1213261 - Ma F, Zhu W, Guan Y, Yang L, Xia X, Chen S, Li Q, Guan X, Yi Z, Qian H, Yi X, Xu B. ctDNA dynamics: a novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget 7(40), 66020-66031 (2016).
https://doi.org/10.18632/oncotarget.11791 - Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA, Jr. Circulating mutant DNA to assess tumor dynamics. Nat Med 14(9), 985-990 (2008).
https://doi.org/10.1038/nm.1789 - Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, Kosmider S, Skinner I, Wong R, Steel M, Tran B, Desai J, Jones I, Haydon A, Hayes T, Price TJ, Strausberg RL, Diaz LA, Jr., Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 8(346), 346ra392 (2016).
https://doi.org/10.1126/scitranslmed.aaf6219 - Weber B, Meldgaard P, Hager H, Wu L, Wei W, Tsai J, Khalil A, Nexo E, Sorensen BS. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 14(1), 294 (2014).
https://doi.org/10.1186/1471-2407-14-294 - Epigenomics receives FDA approval for Epi proColon (2016). Available at https://epigenomics.com/epigenomics-receives-fda-approval-epi-procolon/
- Bauden M, Pamart D, Ansari D, Herzog M, Eccleston M, Micallef J, Andersson B, Andersson R. Circulating nucleosomes as epigenetic biomarkers in pancreatic cancer. Clin Epigenetics 7(1), 106-112 (2015).
https://doi.org/10.1186/s13148-015-0139-4 - Galanopoulos M, Tsoukalas N, Papanikolaou IS, Tolia M, Gazouli M, Mantzaris GJ. Abnormal DNA methylation as a cell-free circulating DNA biomarker for colorectal cancer detection: A review of literature. World J Gastrointest Oncol 9(4), 142-152 (2017).
https://doi.org/10.4251/wjgo.v9.i4.142 - Lissa D, Robles AI. Methylation analyses in liquid biopsy. Transl Lung Cancer Res 5(5), 492-504 (2016).
https://doi.org/10.21037/tlcr.2016.10.03 - Webb S. The cancer bloodhounds. Nat Biotechnol 34(11), 1090-1094 (2016).
https://doi.org/10.1038/nbt.3717 - Han X, Wang J, Sun Y. Circulating Tumor DNA as Biomarkers for Cancer Detection. Genomics Proteomics Bioinformatics 15(2), 59-72 (2017).
https://doi.org/10.1016/j.gpb.2016.12.004 - Jenkins S, Yang JC, Ramalingam SS, Yu K, Patel S, Weston S, Hodge R, Cantarini M, Janne PA, Mitsudomi T, Goss GD. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 12(7), 1061-1070 (2017).
https://doi.org/10.1016/j.jtho.2017.04.003 - Beaver JA, Jelovac D, Balukrishna S, Cochran R, Croessmann S, Zabransky DJ, Wong HY, Toro PV, Cidado J, Blair BG, Chu D, Burns T, Higgins MJ, Stearns V, Jacobs L, Habibi M, Lange J, Hurley PJ, Lauring J, Vandenberg D, Kessler J, Jeter S, Samuels ML, Maar D, Cope L, Cimino-Mathews A, Argani P, Wolff AC, Park BH. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 20(10), 2643-2650 (2014).
https://doi.org/10.1158/1078-0432.CCR-13-2933 - Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in Liquid Biopsy – Current Status and Where We Need to Progress. Comput Struct Biotechnol J 16(1), 190-195 (2018).
https://doi.org/10.1016/j.csbj.2018.05.002 - Bidard FC, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol 15(4), 406-414 (2014).
https://doi.org/10.1016/S1470-2045(14)70069-5 - Bayarri-Lara C, Ortega FG, Cueto Ladron De Guevara A, Puche JL, Ruiz Zafra J, De Miguel-Perez D, Ramos AS, Giraldo-Ospina CF, Navajas Gomez JA, Delgado-Rodriguez M, Lorente JA, Serrano MJ. Circulating Tumor Cells Identify Early Recurrence in Patients with Non-Small Cell Lung Cancer Undergoing Radical Resection. PLoS One 11(2), e0148659 (2016).
https://doi.org/10.1371/journal.pone.0148659 - Vlaeminck-Guillem V. When Prostate Cancer Circulates in the Bloodstream. Diagnostics (Basel) 5(4), 428-474 (2015).
https://doi.org/10.3390/diagnostics5040428 - Huang X, Gao P, Song Y, Sun J, Chen X, Zhao J, Xu H, Wang Z. Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch System in colorectal cancer. BMC Cancer 15(1), 202 (2015).
https://doi.org/10.1186/s12885-015-1218-9 - Ankeny JS, Court CM, Hou S, Li Q, Song M, Wu D, Chen JF, Lee T, Lin M, Sho S, Rochefort MM, Girgis MD, Yao J, Wainberg ZA, Muthusamy VR, Watson RR, Donahue TR, Hines OJ, Reber HA, Graeber TG, Tseng HR, Tomlinson JS. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J Cancer 114(12), 1367-1375 (2016).
https://doi.org/10.1038/bjc.2016.121 - Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351(8), 781-791 (2004).
https://doi.org/10.1056/NEJMoa040766 - Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 26(19), 3213-3221 (2008).
https://doi.org/10.1200/JCO.2007.15.8923 - De Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 14(19), 6302-6309 (2008).
https://doi.org/10.1158/1078-0432.CCR-08-0872 - Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, Scher HI. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res 13(23), 7053-7058 (2007).
https://doi.org/10.1158/1078-0432.CCR-07-1506 - Maheswaran S, Sequist LV, Nagrath S et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 359(4), 366-377 (2008).
https://doi.org/10.1056/NEJMoa0800668 - Markou A, Farkona S, Schiza C, Efstathiou T, Kounelis S, Malamos N, Georgoulias V, Lianidou E. PIK3CA mutational status in circulating tumor cells can change during disease recurrence or progression in patients with breast cancer. Clin Cancer Res 20(22), 5823-5834 (2014).
https://doi.org/10.1158/1078-0432.CCR-14-0149 - Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, Zhao J, Xu L, An T, Ma Q, Wang Y, Wu M, Sun Y, Wang S, Li Z, Yang X, Yong J, Su XD, Lu Y, Bai F, Xie XS, Wang J. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A 110(52), 21083-21088 (2013).
https://doi.org/10.1073/pnas.1320659110 - Chimonidou M, Strati A, Tzitzira A, Sotiropoulou G, Malamos N, Georgoulias V, Lianidou ES. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin Chem 57(8), 1169-1177 (2011).
https://doi.org/10.1373/clinchem.2011.165902 - Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, Johnson A, Jendrisak A, Bambury R, Danila D, Mclaughlin B, Wahl J, Greene SB, Heller G, Marrinucci D, Fleisher M, Dittamore R. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol 2(11), 1441-1449 (2016).
https://doi.org/10.1001/jamaoncol.2016.1828 - Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, Smas ME, Lord JB, Brannigan BW, Trautwein J, Bander NH, Wu CL, Sequist LV, Smith MR, Ramaswamy S, Toner M, Maheswaran S, Haber DA. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov 2(11), 995-1003 (2012).
https://doi.org/10.1158/2159-8290.CD-12-0222 - Guibert N, Delaunay M, Lusque A, Boubekeur N, Rouquette I, Clermont E, Mourlanette J, Gouin S, Dormoy I, Favre G, Mazieres J, Pradines A. PD-L1 expression in circulating tumor cells of advanced non-small cell lung cancer patients treated with nivolumab. Lung Cancer 120(1), 108-112 (2018).
https://doi.org/10.1016/j.lungcan.2018.04.001 - Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. Isolation by size of epithelial tumor cells : a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol 156(1), 57-63 (2000).
https://doi.org/10.1016/S0002-9440(10)64706-2 - Li P, Stratton ZS, Dao M, Ritz J, Huang TJ. Probing circulating tumor cells in microfluidics. Lab Chip 13(4), 602-609 (2013).
https://doi.org/10.1039/c2lc90148j - Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J, Saada V, Farace F, Vielh P, Malaquin L, Viovy JL. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. Proc Natl Acad Sci USA 107(33), 14524-14529 (2010).
https://doi.org/10.1073/pnas.1001515107 - Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol 192(3), 373-382 (2011).
https://doi.org/10.1083/jcb.201010021 - Kallergi G, Politaki E, Alkahtani S, Stournaras C, Georgoulias V. Evaluation of Isolation Methods for Circulating Tumor Cells (CTCs). Cell Physiol Biochem 40(3-4), 411-419 (2016).
https://doi.org/10.1159/000452556 - Marsavela G, Aya-Bonilla CA, Warkiani ME, Gray ES, Ziman M. Melanoma circulating tumor cells: Benefits and challenges required for clinical application. Cancer Lett 424(1), 1-8 (2018).
https://doi.org/10.1016/j.canlet.2018.03.013 - Wu S, Liu S, Liu Z, Huang J, Pu X, Li J, Yang D, Deng H, Yang N, Xu J. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS One 10(4), e0123976 (2015).
https://doi.org/10.1371/journal.pone.0123976 - Gradilone A, Raimondi C, Nicolazzo C, Petracca A, Gandini O, Vincenzi B, Naso G, Agliano AM, Cortesi E, Gazzaniga P. Circulating tumour cells lacking cytokeratin in breast cancer: the importance of being mesenchymal. J Cell Mol Med 15(5), 1066-1070 (2011).
https://doi.org/10.1111/j.1582-4934.2011.01285.x - Kallergi G, Papadaki MA, Politaki E, Mavroudis D, Georgoulias V, Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res 13(3), R59 (2011).
https://doi.org/10.1186/bcr2896 - Gorges TM, Tinhofer I, Drosch M, Rose L, Zollner TM, Krahn T, Von Ahsen O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 12(1), 178 (2012).
https://doi.org/10.1186/1471-2407-12-178 - Wu F, Zhu J, Mao Y, Li X, Hu B, Zhang D. Associations between the Epithelial-Mesenchymal Transition Phenotypes of Circulating Tumor Cells and the Clinicopathological Features of Patients with Colorectal Cancer. Dis Markers 2017(9474532), 1-6 (2017).
https://doi.org/10.1155/2017/9474532 - Loh J, Jovanovic L, Lehman M, Capp A, Pryor D, Harris M, Nelson C, Martin J. Circulating tumor cell detection in high-risk non-metastatic prostate cancer. J Cancer Res Clin Oncol 140(12), 2157-2162 (2014).
https://doi.org/10.1007/s00432-014-1775-3 - Andree KC, Van Dalum G, Terstappen LW. Challenges in circulating tumor cell detection by the CellSearch system. Mol Oncol 10(3), 395-407 (2016).
https://doi.org/10.1016/j.molonc.2015.12.002 - Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Flejou JF, Lantuejoul S, Piaton E, Mourad N, Butori C, Selva E, Poudenx M, Sibon S, Kelhef S, Venissac N, Jais JP, Mouroux J, Molina TJ, Hofman P. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 17(4), 827-835 (2011).
https://doi.org/10.1158/1078-0432.CCR-10-0445 - Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr., Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A 107(43), 18392-18397 (2010).
https://doi.org/10.1073/pnas.1012539107 - Van Der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 64(3), 676-705 (2012).
https://doi.org/10.1124/pr.112.005983 - Harding C, Stahl P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem Biophys Res Commun 113(2), 650-658 (1983).
https://doi.org/10.1016/0006-291X(83)91776-X - Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, Azab F, Flores LM, Campigotto F, Weller E, Anderson KC, Scadden DT, Ghobrial IM. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest 123(4), 1542-1555 (2013).
https://doi.org/10.1172/JCI66517 - Melo SA, Sugimoto H, O’connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26(5), 707-721 (2014).
https://doi.org/10.1016/j.ccell.2014.09.005 - Svensson KJ, Kucharzewska P, Christianson HC, Skold S, Lofstedt T, Johansson MC, Morgelin M, Bengzon J, Ruf W, Belting M. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc Natl Acad Sci USA 108(32), 13147-13152 (2011).
https://doi.org/10.1073/pnas.1104261108 - Syn N, Wang L, Sethi G, Thiery JP, Goh BC. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci 37(7), 606-617 (2016).
https://doi.org/10.1016/j.tips.2016.04.006 - Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, Howell SB. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther 4(10), 1595-1604 (2005).
https://doi.org/10.1158/1535-7163.MCT-05-0102 - Ciravolo V, Huber V, Ghedini GC, Venturelli E, Bianchi F, Campiglio M, Morelli D, Villa A, Della Mina P, Menard S, Filipazzi P, Rivoltini L, Tagliabue E, Pupa SM. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol 227(2), 658-667 (2012).
https://doi.org/10.1002/jcp.22773 - Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 10(1), 42-46 (2009).
https://doi.org/10.3816/CLC.2009.n.006 - Khan S, Bennit HF, Turay D, Perez M, Mirshahidi S, Yuan Y, Wall NR. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 14(1), 176 (2014).
https://doi.org/10.1186/1471-2407-14-176 - Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, Lebleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523(7559), 177-182 (2015).
https://doi.org/10.1038/nature14581 - Skog J, Wurdinger T, Van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr., Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10(12), 1470-1476 (2008).
https://doi.org/10.1038/ncb1800 - Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110(1), 13-21 (2008).
https://doi.org/10.1016/j.ygyno.2008.04.033 - Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, Blelloch R. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell 177(2), 414-427 e413 (2019).
https://doi.org/10.1016/j.cell.2019.02.016 - Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, Mcgettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560(7718), 382-386 (2018).
https://doi.org/10.1038/s41586-018-0392-8 - Coumans FaW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S, Emanueli C, Gasecka A, Hendrix A, Hill AF, Lacroix R, Lee Y, Van Leeuwen TG, Mackman N, Mager I, Nolan JP, Van Der Pol E, Pegtel DM, Sahoo S, Siljander PRM, Sturk G, De Wever O, Nieuwland R. Methodological Guidelines to Study Extracellular Vesicles. Circ Res 120(10), 1632-1648 (2017).
https://doi.org/10.1161/CIRCRESAHA.117.309417 - Kilgour E, Rothwell DG, Brady G, Dive C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 37(4), 485-495 (2020).
https://doi.org/10.1016/j.ccell.2020.03.012 - Zhang Z, Xiao Y, Zhao J, Chen M, Xu Y, Zhong W, Xing J, Wang M. Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology 21(3), 519-525 (2016).
https://doi.org/10.1111/resp.12696 - Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14(1), 319-332 (2013).
https://doi.org/10.1186/1471-2164-14-319 - Takahashi Y, Nishikawa M, Shinotsuka H, Matsui Y, Ohara S, Imai T, Takakura Y. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol 165(2), 77-84 (2013).
https://doi.org/10.1016/j.jbiotec.2013.03.013 - Mentkowski KI, Snitzer JD, Rusnak S, Lang JK. Therapeutic Potential of Engineered Extracellular Vesicles. AAPS J 20(3), 50 (2018).
https://doi.org/10.1208/s12248-018-0211-z - Giordano A, Giuliano M, De Laurentiis M, Arpino G, Jackson S, Handy BC, Ueno NT, Andreopoulou E, Alvarez RH, Valero V, De Placido S, Hortobagyi GN, Reuben JM, Cristofanilli M. Circulating tumor cells in immunohistochemical subtypes of metastatic breast cancer: lack of prediction in HER2-positive disease treated with targeted therapy. Ann Oncol 23(5), 1144-1150 (2012).
https://doi.org/10.1093/annonc/mdr434 - Coumans FA, Ligthart ST, Uhr JW, Terstappen LW. Challenges in the enumeration and phenotyping of CTC. Clin Cancer Res 18(20), 5711-5718 (2012).
https://doi.org/10.1158/1078-0432.CCR-12-1585 - Bork U, Rahbari NN, Scholch S, Reissfelder C, Kahlert C, Buchler MW, Weitz J, Koch M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: a prospective study. Br J Cancer 112(8), 1306-1313 (2015).
https://doi.org/10.1038/bjc.2015.88 - Li Z, Ma YY, Wang J, Zeng XF, Li R, Kang W, Hao XK. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther 9(1), 139-148 (2016).
https://doi.org/10.2147/OTT.S95565 - Kulasinghe A, Kapeleris J, Kimberley R, Mattarollo SR, Thompson EW, Thiery JP, Kenny L, O’byrne K, Punyadeera C. The prognostic significance of circulating tumor cells in head and neck and non-small-cell lung cancer. Cancer Med 7(12), 5910-5919 (2018).
https://doi.org/10.1002/cam4.1832 - Li Y, Tian X, Gao L, Jiang X, Fu R, Zhang T, Ren T, Hu P, Wu Y, Zhao P, Yang D. Clinical significance of circulating tumor cells and tumor markers in the diagnosis of lung cancer. Cancer Med 8(8), 3782-3792 (2019).
https://doi.org/10.1002/cam4.2286 - Hong X, Sullivan RJ, Kalinich M, Kwan TT, Giobbie-Hurder A, Pan S, Licausi JA, Milner JD, Nieman LT, Wittner BS, Ho U, Chen T, Kapur R, Lawrence DP, Flaherty KT, Sequist LV, Ramaswamy S, Miyamoto DT, Lawrence M, Toner M, Isselbacher KJ, Maheswaran S, Haber DA. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc Natl Acad Sci U S A 115(10), 2467-2472 (2018).
https://doi.org/10.1073/pnas.1719264115 - Gao Y, Ni X, Guo H, Su Z, Ba Y, Tong Z, Guo Z, Yao X, Chen X, Yin J, Yan Z, Guo L, Liu Y, Bai F, Xie XS, Zhang N. Single-cell sequencing deciphers a convergent evolution of copy number alterations from primary to circulating tumor cells. Genome Res 27(8), 1312-1322 (2017).
https://doi.org/10.1101/gr.216788.116 - Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, Krol I, Scheidmann MC, Beisel C, Stirnimann CU, Kurzeder C, Heinzelmann-Schwarz V, Rochlitz C, Weber WP, Aceto N. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 176(1-2), 98-112 e114 (2019).
https://doi.org/10.1016/j.cell.2018.11.046 - Wang HL, Wang SS, Song WH, Pan Y, Yu HP, Si TG, Liu Y, Cui XN, Guo Z. Expression of prostate-specific membrane antigen in lung cancer cells and tumor neovasculature endothelial cells and its clinical significance. PLoS One 10(5), e0125924 (2015).
https://doi.org/10.1371/journal.pone.0125924 - Wallwiener M, Hartkopf AD, Riethdorf S, Nees J, Sprick MR, Schonfisch B, Taran FA, Heil J, Sohn C, Pantel K, Trumpp A, Schneeweiss A. The impact of HER2 phenotype of circulating tumor cells in metastatic breast cancer: a retrospective study in 107 patients. BMC Cancer 15(1), 403-409 (2015).
https://doi.org/10.1186/s12885-015-1423-6 - Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L, Fais S. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One 4(4), e5219 (2009).
https://doi.org/10.1371/journal.pone.0005219 - Khan S, Jutzy JM, Valenzuela MM, Turay D, Aspe JR, Ashok A, Mirshahidi S, Mercola D, Lilly MB, Wall NR. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One 7(10), e46737 (2012).
https://doi.org/10.1371/journal.pone.0046737 - Valenzuela MM, Ferguson Bennit HR, Gonda A, Diaz Osterman CJ, Hibma A, Khan S, Wall NR. Exosomes Secreted from Human Cancer Cell Lines Contain Inhibitors of Apoptosis (IAP). Cancer Microenviron 8(2), 65-73 (2015).
https://doi.org/10.1007/s12307-015-0167-9 - Khan S, Simpson J, Lynch JC, Turay D, Mirshahidi S, Gonda A, Sanchez TW, Casiano CA, Wall NR. Racial differences in the expression of inhibitors of apoptosis (IAP) proteins in extracellular vesicles (EV) from prostate cancer patients. PLoS One 12(10), e0183122 (2017).
https://doi.org/10.1371/journal.pone.0183122 - Smalley DM, Sheman NE, Nelson K, Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res 7(5), 2088-2096 (2008).
https://doi.org/10.1021/pr700775x - Chen CL, Lai YF, Tang P, Chien KY, Yu JS, Tsai CH, Chen HW, Wu CC, Chung T, Hsu CW, Chen CD, Chang YS, Chang PL, Chen YT. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res 11(12), 5611-5629 (2012).
https://doi.org/10.1021/pr3008732 - Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310(5748), 644-648 (2005).
https://doi.org/10.1126/science.1117679 - Corcoran C, Friel AM, Duffy MJ, Crown J, O’driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem 57(1), 18-32 (2011).
https://doi.org/10.1373/clinchem.2010.150730 - Giallombardo M, Chacartegui Borras J, Castiglia M, Van Der Steen N, Mertens I, Pauwels P, Peeters M, Rolfo C. Exosomal miRNA Analysis in Non-small Cell Lung Cancer (NSCLC) Patients’ Plasma Through qPCR: A Feasible Liquid Biopsy Tool. J Vis Exp 1(111), 53900-53905 (2016).
https://doi.org/10.3791/53900 - Zheng G, Du L, Yang X, Zhang X, Wang L, Yang Y, Li J, Wang C. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer 111(10), 1985-1992 (2014).
https://doi.org/10.1038/bjc.2014.489 - Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano Y, Shinden Y, Eguchi H, Yamamoto H, Doki Y, Mori M, Ochiya T, Mimori K. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer 113(2), 275-281 (2015).
https://doi.org/10.1038/bjc.2015.201
All site content, except where otherwise noted, is licensed under a Creative Commons Attribution 4.0 License